Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification, Characterization, and Phylogenetic Analyses of MiOFPs
2.3. Phenotypic and Histological Observations
2.4. RNA-Seq Analysis
2.5. Total RNA Extraction and RT-qPCR Analysis
3. Results
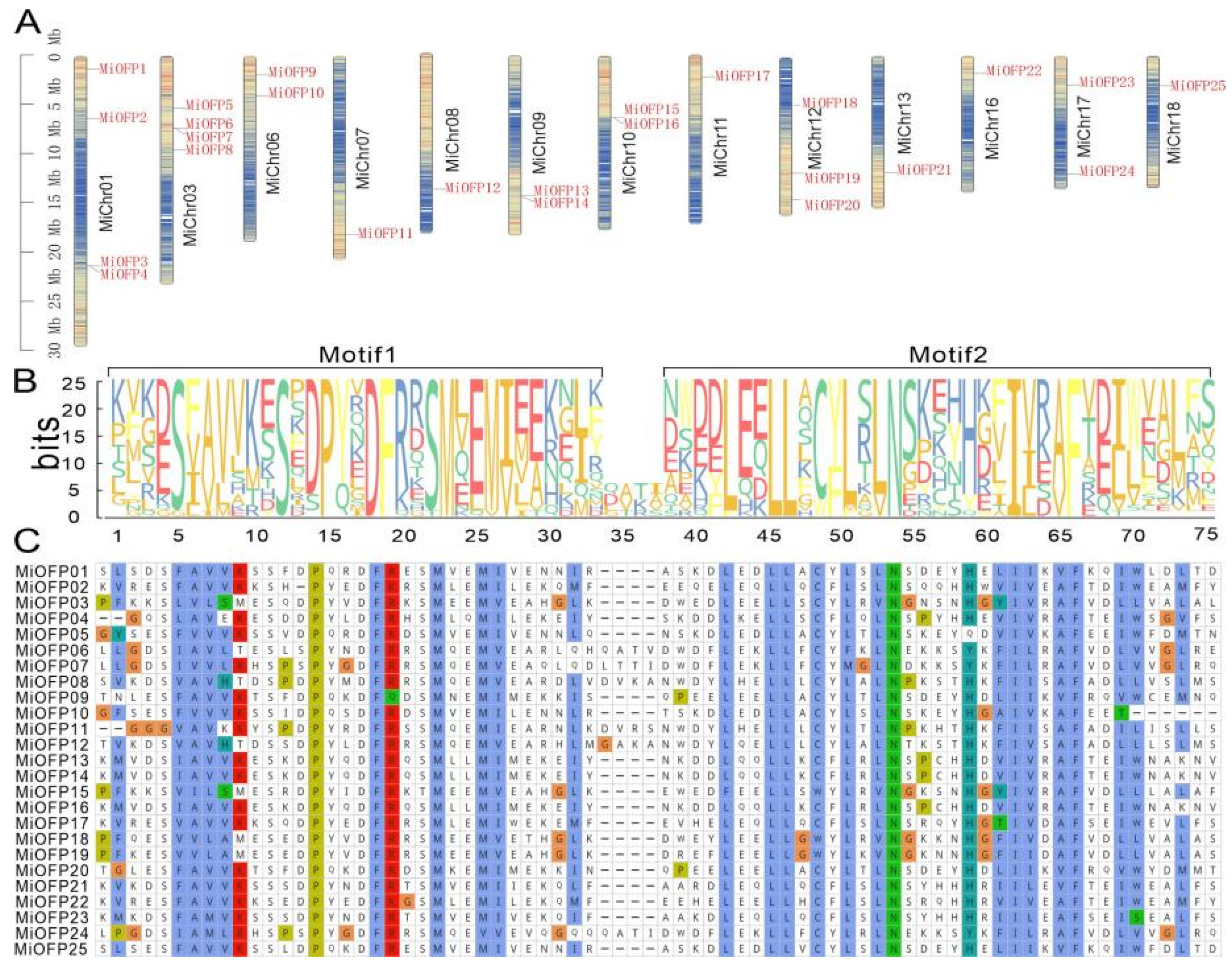
3.1. Identification of OFP Genes in Mango
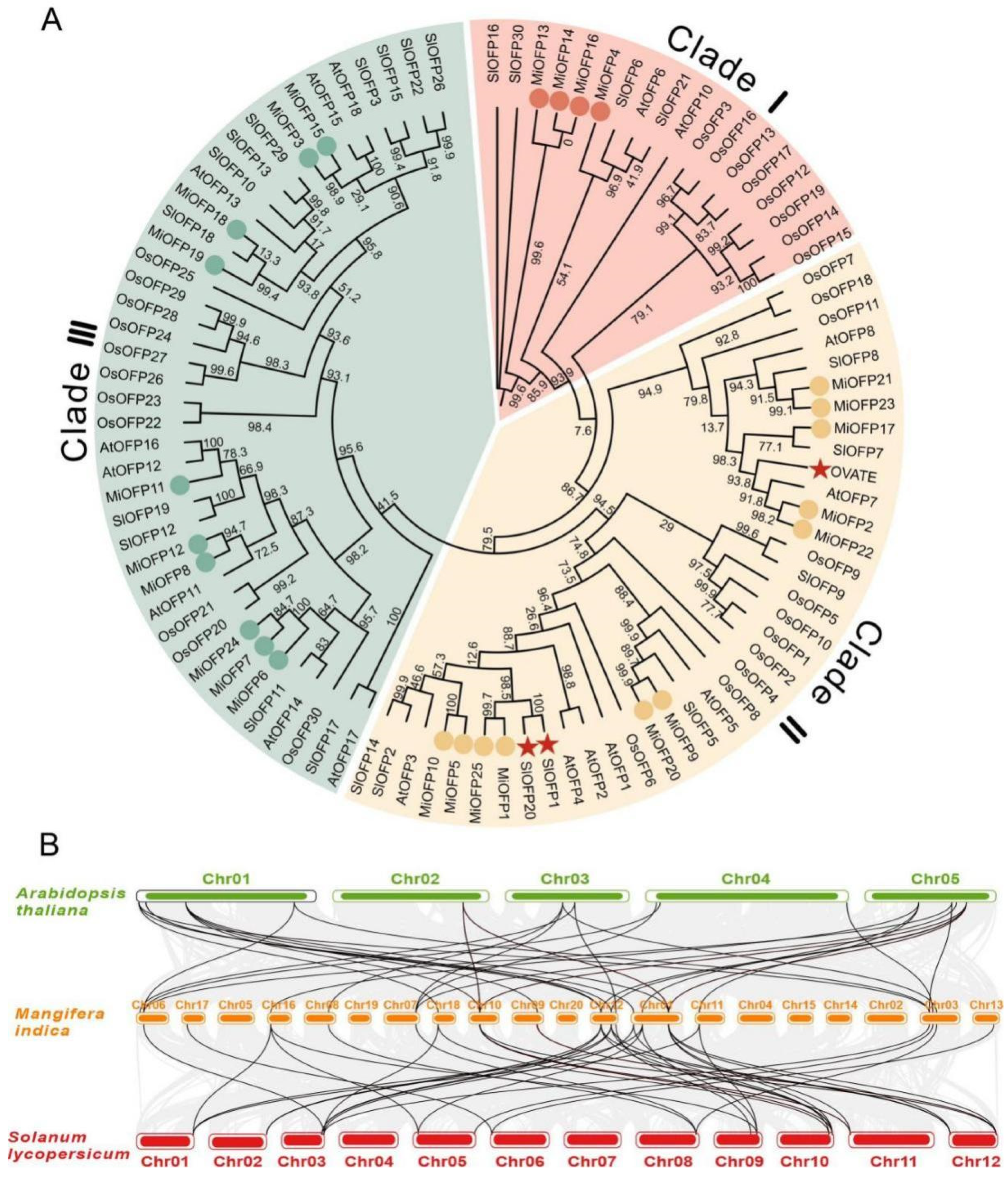
3.2. Sequence Alignment, Structure, and Phylogenetic Analysis of the MiOFPs
3.3. Expression Analysis of MiOFP Genes in Different Tissues
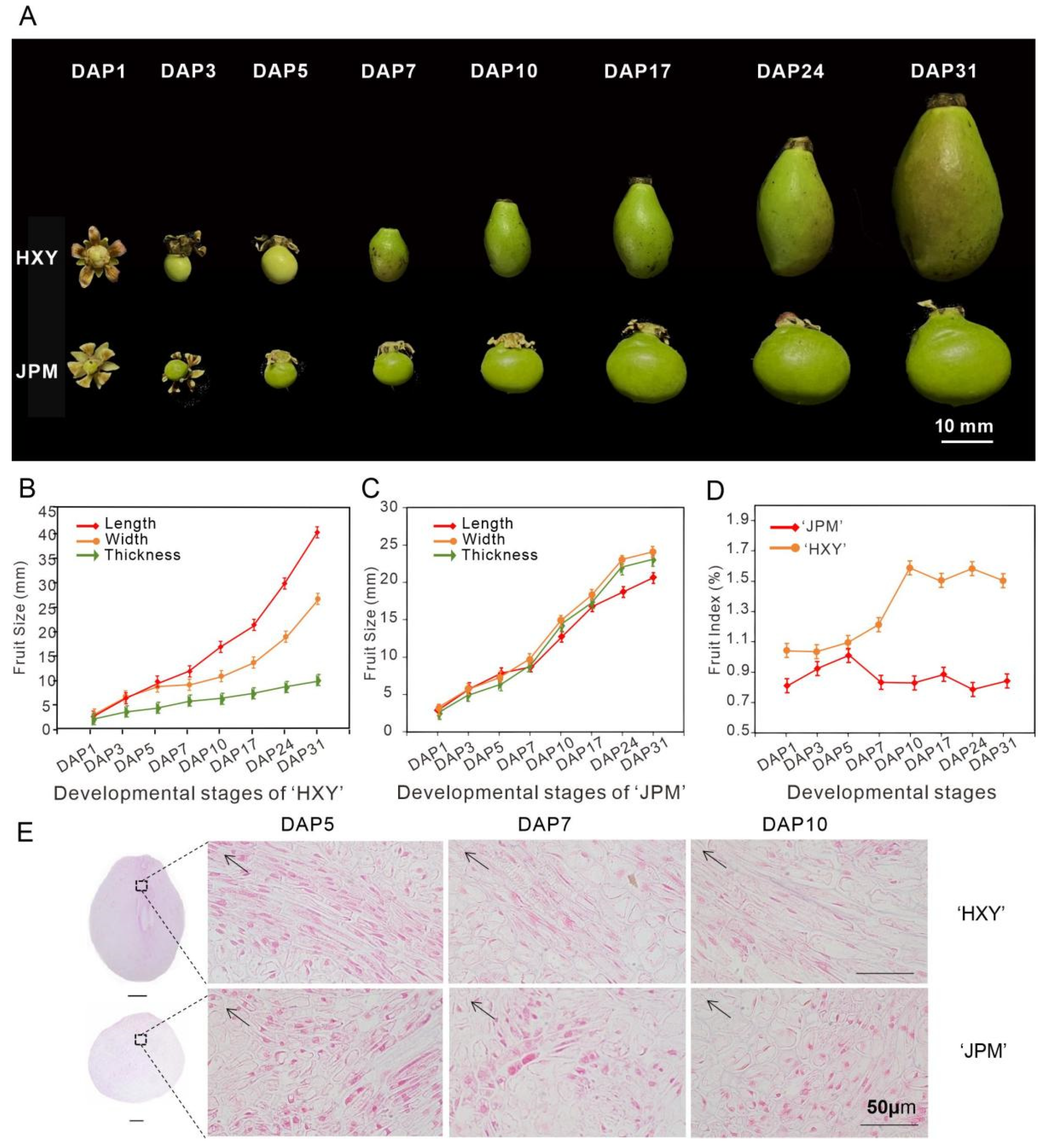
3.4. Morphological Analysis of Fruit Development
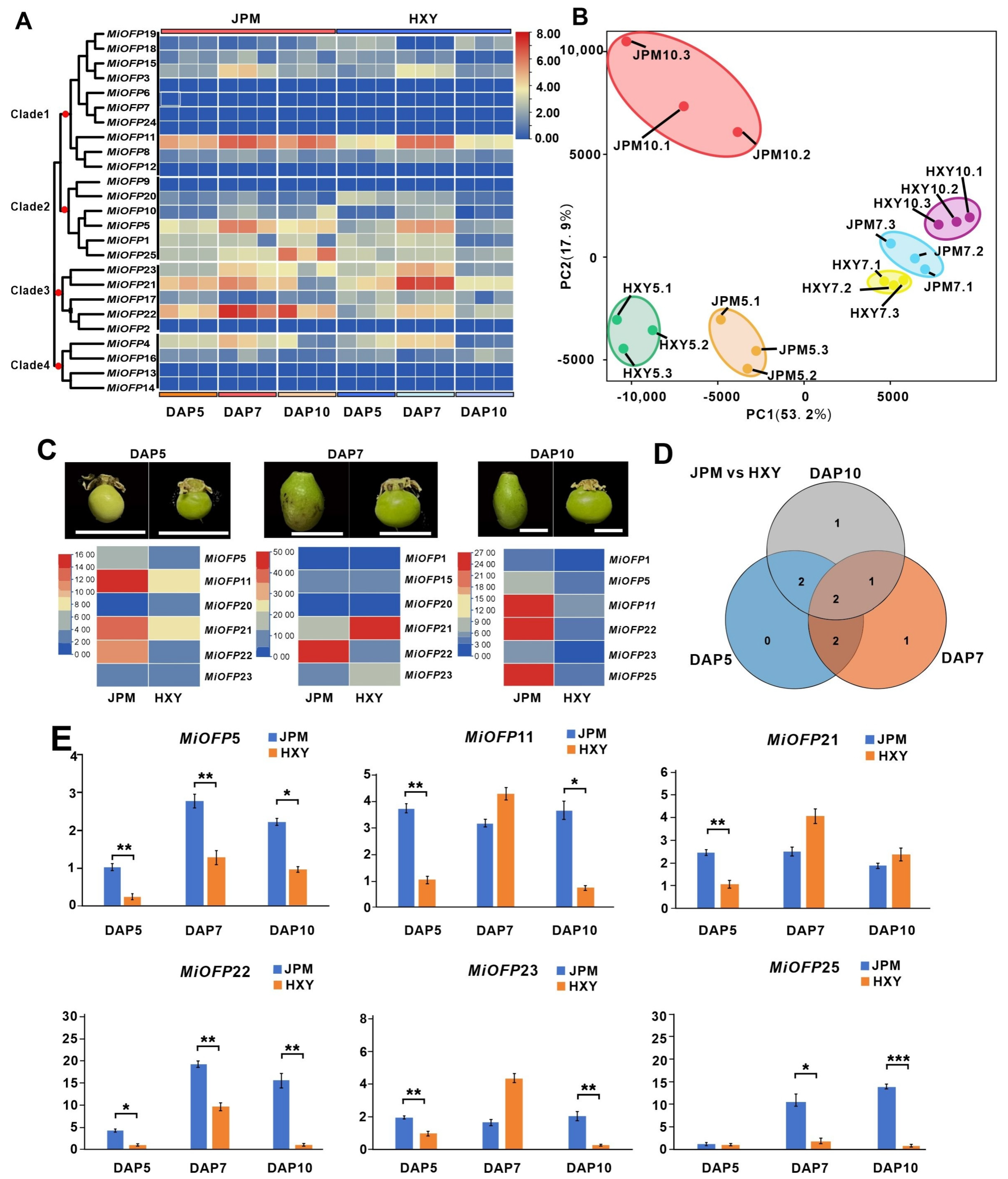
3.5. Expression Profile of MiOFP Genes in Two Varieties with Contrasting Fruit Shape
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, C.; Xie, D.; Bai, T.; Luo, X.; Zhang, F.; Ni, Z.; Chen, Y. Diversity of a Large Collection of Natural Populations of Mango (Mangifera indica Linn.) Revealed by Agro-Morphological and Quality Traits. Diversity 2020, 12, 27. [Google Scholar] [CrossRef]
- Kulkarni, M.M.; Burondkar, M.M.; Dalvi, N.V.; Salvi, B.R.; Haldankar, P.M.; Bhattacharyya, T. Mango Fruit Size Diversity found in Konkan. Adv. Agric. Res. Technol. J. 2019, 3, 43–46. [Google Scholar]
- Zhu, Q.; Deng, L.; Chen, J.; Rodríguez, G.R.; Sun, C.; Chang, Z.; Yang, T.; Zhai, H.; Jiang, H.; Topcu, Y.; et al. Redesigning the tomato fruit shape for mechanized production. Nat. Plants 2023, 9, 1659–1674. [Google Scholar] [CrossRef] [PubMed]
- van der Knaap, E.; Tanksley, S.D. The making of a bell pepper-shaped tomato fruit: Identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theor. Appl. Genet. 2003, 107, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ma, R.J.; Gao, L.; Zhang, J.; Zhang, A.; Zhang, X.; Ren, F.; Zhang, W.; Liao, L.; Yang, Q.; et al. A 1.7-Mb chromosomal inversion downstream of a PpOFP1 gene is responsible for flat fruit shape in peach. Plant Biotechnol. J. 2021, 19, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Chen, Y.; Luo, R.; Huang, J.; Zhao, Z.; Wang, W.; Wang, Y.; Dang, Z. Development Status of Chinese Mango Industry in 2018. Adv. Agric. Hortic. Entomol. 2019, 2019, 1–6. [Google Scholar]
- Liu, J.; Joyce, V.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef] [PubMed]
- Gui, B.; Wang, Y. Cloning and Sequence Analysis of ovate Orthologous Gene in Tobacco (Nicotiana tabacum L.). Plant Physiol. Commun. 2007, 43, 1050–1056. [Google Scholar]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the Tomato Germplasm and the Relationship to Fruit Shape Diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef]
- Tsaballa, A.; Pasentsis, K.; Darzentas, N.; Tsaftaris, A.S. Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 2011, 11, 46. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Guo, J.; Zeng, Q.; Ellis, B.; Chen, J. Arabidopsis Ovate Family Proteins, a Novel Transcriptional Repressor Family, Control Multiple Aspects of Plant Growth and Development. PLoS ONE 2011, 6, e23896. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Van, H.; Gonzalez, G.; Xiao, H.; van der Knaap, E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 2013, 288, 111–129. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Guo, J.; Chen, J. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007, 50, 858–872. [Google Scholar] [CrossRef]
- Schmitz, A.J.; Begcy, K.; Sarath, G.; Walia, H. Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 2015, 241, 177–188. [Google Scholar] [CrossRef]
- Yang, C.; Ma, Y.; He, Y.; Tian, Z.; Li, J. OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J. 2018, 93, 489–501. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodriguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef] [PubMed]
- Snouffer, A.; Kraus, C.; van der Knaap, E. The shape of things to come: Ovate family proteins regulate plant organ shape. Curr. Opin. Plant Biol. 2020, 53, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Hao, W.; Sun, H.; Zhang, L. Characterization of the OFP Gene Family and its Putative Involvement of Tuberous Root Shape in Radish. Int. J. Mol. Sci. 2020, 21, 1293. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Sun, W.; Yuan, Y.; Zhang, N.; Hayward, A.; Liu, Y.; Wang, Y. Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann. Bot. 2014, 113, 1219–1233. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jiang, W.; Liu, Q.; Zhang, H.; Piao, M.; Chen, Z. Expression Pattern and Subcellular Localization of the Ovate Protein Family in Rice. PLoS ONE 2015, 10, e0118966. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, Y.; Gao, S.; Tao, J. Bioinformatics and Expression of the OVATE Gene Family in Grape. Sci. Agric. Sin. 2016, 49, 3787–3797. [Google Scholar] [CrossRef]
- Qi, W.; Luo, Z.; Li, H.; Zhang, Z.; Yan, C.; Wang, J.; Liu, M. Identification and Bioinformatics Analysis of OVATE Gene Family in Chinese Jujube. Mol. Plant Breed. 2021, 19, 2207–2216. [Google Scholar]
- Li, H.; Dong, Q.; Zhao, Q.; Ran, K. Genome-wide identification, expression profiling, and protein-protein interaction properties of ovate family proteins in apple. Tree Genet. Genomes 2019, 15, 45. [Google Scholar] [CrossRef]
- Wang, P.; Luo, Y.; Huang, J.; Gao, S.; Zhu, G.; Dang, Z.; Gai, J.; Yang, M.; Zhu, M.; Zhang, H.; et al. The genome evolution and domestication of tropical fruit mango. Genome Biol. 2020, 21, 60. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ada, T.F.; Delia, W. Tissue Processing and Hematoxylin and Eosin Staining. Histopathol. Methods Protoc. Methods Mol. Biol. 2014, 1180, 31–43. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 22−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gao, H.; Li, P.; Donf, T. Advances in the Study off Actors Influencing Fruit Shape. J. Trop. Biol. 2016, 7, 280–284. [Google Scholar] [CrossRef]
- Guo, J.; Cao, K.; Li, Y.; Yao, J.; Deng, C.; Wang, Q.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; et al. Comparative Transcriptome and Microscopy Analyses Provide Insights into Flat Shape Formation in Peach (Prunus persica). Front. Plant Sci. 2018, 8, 2215. [Google Scholar] [CrossRef]
- Gillaspy, G.; Hilla, B.D.; Wilhelm, G. Fruits: A Developmental Perspective. Plant Cell 1993, 5, 1439–1451. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Ellis, B. Overview of OVATE FAMILY PROTEINS, A Novel Class of Plant-Specific Growth Regulators. Front. Plant Sci. 2016, 7, 417. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Zhu, J.W.; Lao, A.; Li, Y.Z.; Xia, L.M.; MO, X.; Hu, W.L.; He, X.H.; Luo, C. Expression and functional analysis of MiOFP1 gene in mango. J. Fruit Sci. 2023, 40, 1099–1108. [Google Scholar] [CrossRef]
- Bleckmann, A.; Weidtkamp-Peters, S.; Seidel, C.A.M.; Simon, R. Stem Cell Signaling in Arabidopsis Requires CRN to Localize CLV2 to the Plasma Membrane. Plant Physiol. 2010, 152, 166–176. [Google Scholar] [CrossRef]
- Mauxion, J.P.; Chevalier, C.; Gonzalez, N. Complex cellular and molecular events determining fruit size. Trends Plant Sci. 2021, 26, 1023–1038. [Google Scholar] [CrossRef]
- Chen, H.B.; Huang, M.Y. Fruit growth and abscission of the mango (Mangifera indica L.) ZiHua. J. South China Agric. Univ. 1995, 16, 73–77. [Google Scholar]
- Xu, X.; Wang, X.; Zhou, S.; Huang, X.; Liu, P.; Ma, B.; Chen, X. Genome-Wide Identification and Characterization of the OFP Gene Family in the Wild Strawberry Fragaria vesca. Agronomy 2024, 14, 569. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, S.; Luo, X.; Li, J.; Li, T.; Tang, X.; Liu, F.; Zou, X.; Qin, C. Genomewide analysis of OFP gene family in pepper (Capsicum annuum L.). Front. Genet. 2022, 13, 941–954. [Google Scholar] [CrossRef] [PubMed]


| Primer | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Amplicon Size |
|---|---|---|---|
| MiOFP5 | TCCGAAAGTTGTGGTCATCGA | ACCACCACAAAGCTCTCCG | 342 |
| MiOFP11 | TCGCCACCATTTTCGCCT | CCCGCCTCCTTTGAGTGG | 227 |
| MiOFP21 | ACAGCACCACCGTTACCG | TCACCACCGCAAAGCTGT | 251 |
| MiOFP22 | CCCTCCACTCTCCCGTCA | ATCGCGGAGGAAACGTGG | 357 |
| MiOFP23 | GATGCCCAGAAGCCAGCA | CGATATGGGCGAGGCAGG | 246 |
| MiOFP25 | TAGCTGCCGTGCCACAAT | AGAGTCACCGGCAGCAAC | 391 |
| MiActin | ATCTGCTGGAAGGTGCTGAG | CCAAGCAGCATGAAGATCAA | 377 |
| Gene Name | Gene ID | Chromosome Location | Size/aa | Molecular Weight/Da | pI | Hydro- Pathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| MiOFP1 | LOC123225041 | Chr01:1286519–1287906 | 392 | 44,485.46 | 9.55 | −0.744 | Chloroplast |
| MiOFP2 | LOC123195071 | Chr01:6284252–6285495 | 356 | 40,684.17 | 9.47 | −0.658 | Chloroplast |
| MiOFP3 | LOC123193314 | Chr01:21257006–21258100 | 280 | 30,946.09 | 4.99 | −0.542 | Nucleus |
| MiOFP4 | LOC123192493 | Chr01:21271282–21272056 | 186 | 20,802.65 | 9.66 | −0.594 | Chloroplast |
| MiOFP5 | LOC123211132 | Chr03:5224343–5225845 | 342 | 39,396.61 | 9.33 | −0.82 | Chloroplast |
| MiOFP6 | LOC123210957 | Chr03:7301842–7302579 | 245 | 28,217.85 | 6.96 | −0.66 | Chloroplast |
| MiOFP7 | LOC123210960 | Chr03:7306821–7311155 | 246 | 27,779.89 | 4.72 | −0.496 | Nucleus |
| MiOFP8 | LOC123212208 | Chr03:9478321–9479265 | 234 | 26,334.21 | 4.85 | −0.468 | Chloroplast |
| MiOFP9 | LOC123219021 | Chr06:1870688–1872202 | 411 | 47,439.15 | 9.41 | −0.944 | Chloroplast |
| MiOFP10 | LOC123219408 | Chr06:3989550–3990915 | 317 | 35,881.41 | 9.71 | −0.874 | Chloroplast |
| MiOFP11 | LOC123221257 | Chr07:18070119–18071499 | 227 | 25,157.93 | 5.46 | −0.512 | Nucleus |
| MiOFP12 | LOC123223125 | Chr08:13809269–13810321 | 234 | 26,385.33 | 5.68 | −0.476 | Chloroplast |
| MiOFP13 | LOC123226435 | Chr09:14142901–14143877 | 183 | 20,445.09 | 6.08 | −0.652 | Chloroplast |
| MiOFP14 | LOC123226449 | Chr09:14365554–14366530 | 183 | 20,445.09 | 6.08 | −0.652 | Chloroplast |
| MiOFP15 | LOC123228213 | Chr10:6211065–6212266 | 288 | 31,890.43 | 4.94 | −0.453 | Nucleus |
| MiOFP16 | LOC123228223 | Chr10:6224341–6225119 | 178 | 20,108.71 | 9.37 | −0.626 | Chloroplast |
| MiOFP17 | LOC123230016 | Chr11:2215612–2216944 | 318 | 36,541.54 | 9.54 | −0.828 | Mitochondrion |
| MiOFP18 | LOC123192669 | Chr12:5878219–5879439 | 239 | 26,570.41 | 4.62 | −0.339 | Nucleus |
| MiOFP19 | LOC123193551 | Chr12:11701597–11702637 | 235 | 26,093.09 | 4.92 | −0.298 | Nucleus |
| MiOFP20 | LOC123192568 | Chr12:14354392–14355878 | 394 | 45,728.22 | 9.52 | −0.962 | Chloroplast |
| MiOFP21 | LOC123194383 | Chr13:11784106–11785381 | 303 | 35,157.89 | 9.67 | −0.795 | Chloroplast |
| MiOFP22 | LOC123199657 | Chr16:1741505–1742791 | 357 | 40,800.53 | 9.43 | −0.642 | Chloroplast |
| MiOFP23 | LOC123200363 | Chr17:2891367–2892514 | 296 | 33,965.45 | 9.53 | −0.758 | Mitochondrion |
| MiOFP24 | LOC123200256 | Chr17:11932352–11933258 | 255 | 28,843.27 | 5.77 | −0.64 | Chloroplast |
| MiOFP25 | LOC123202271 | Chr18:2977275–2978794 | 391 | 44,206.97 | 9.56 | −0.8 | Chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Xia, R.; Yang, J.; Chen, R.; Zeng, Z.; Fan, C. Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango. Genes 2024, 15, 823. https://doi.org/10.3390/genes15070823
Wu Q, Xia R, Yang J, Chen R, Zeng Z, Fan C. Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango. Genes. 2024; 15(7):823. https://doi.org/10.3390/genes15070823
Chicago/Turabian StyleWu, Qiuping, Rui Xia, Jie Yang, Rong Chen, Zaohai Zeng, and Chao Fan. 2024. "Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango" Genes 15, no. 7: 823. https://doi.org/10.3390/genes15070823
APA StyleWu, Q., Xia, R., Yang, J., Chen, R., Zeng, Z., & Fan, C. (2024). Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango. Genes, 15(7), 823. https://doi.org/10.3390/genes15070823





