Clinical Relevance of the Systematic Analysis of Copy Number Variants in the Genetic Study of Cardiomyopathies
Abstract
1. Introduction
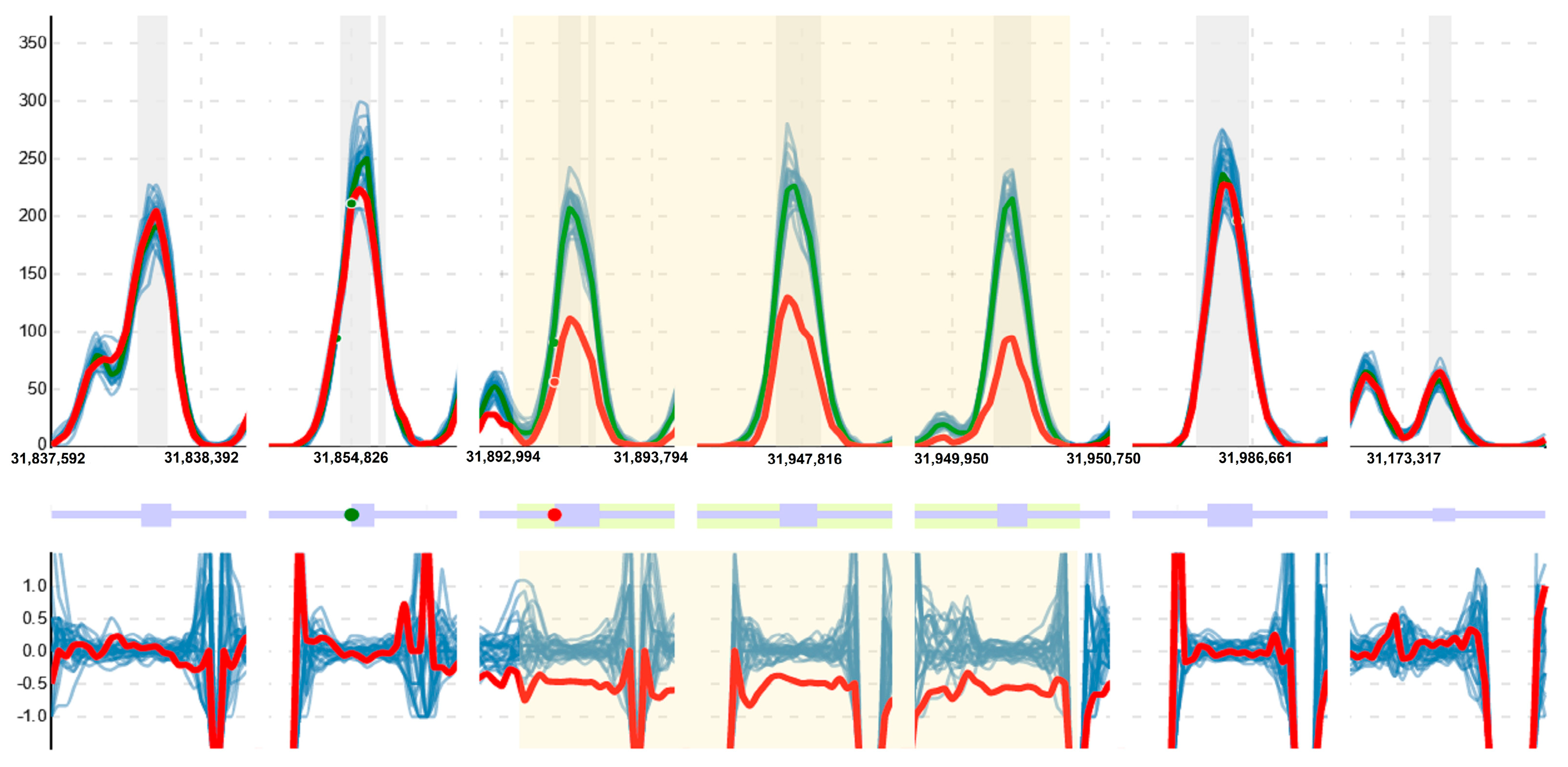
2. Materials and Methods
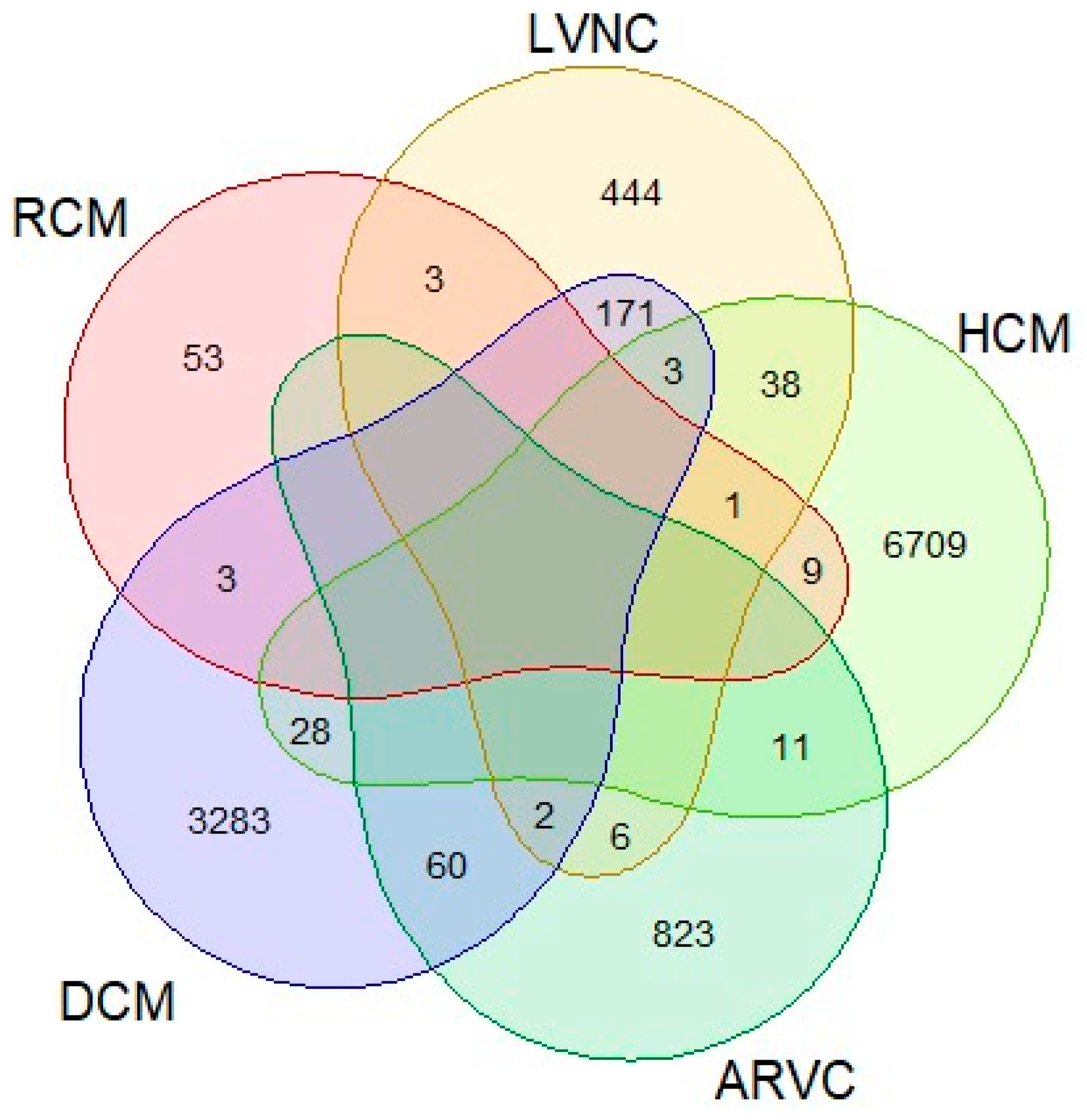
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elliott, P.; Andersson, B.; Arbustini, E.; Bilinska, Z.; Cecchi, F.; Charron, P.; Dubourg, O.; Kuhl, U.; Maisch, B.; McKenna, W.J.; et al. Classification of the Cardiomyopathies: A Position Statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2007, 29, 270–276. [Google Scholar] [CrossRef] [PubMed]
- McKenna, W.J.; Maron, B.J.; Thiene, G. Classification, Epidemiology, and Global Burden of Cardiomyopathies. Circ. Res. 2017, 121, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, J.E.; Hershberger, R.E. Genetic Cardiomyopathies. Curr. Opin. Cardiol. 2018, 33, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, M.J.; Priori, S.G.; Willems, S.; Berul, C.; Brugada, R.; Calkins, H.; Camm, A.J.; Ellinor, P.T.; Gollob, M.; Hamilton, R.; et al. HRS/EHRA Expert Consensus Statement on the State of Genetic Testing for the Channelopathies and Cardiomyopathies. Heart Rhythm 2011, 8, 1308–1339. [Google Scholar] [CrossRef] [PubMed]
- Monserrat, L.; Ortiz-Genga, M.; Lesende, I.; Garcia-Giustiniani, D.; Barriales-Villa, R.; Una-Iglesias, D.; Syrris, P.; Castro-Beiras, A. Genetics of Cardiomyopathies: Novel Perspectives with Next Generation Sequencing. CPD 2014, 21, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Arbelo, E.; Protonotarios, A.; Gimeno, J.R.; Arbustini, E.; Barriales-Villa, R.; Basso, C.; Bezzina, C.R.; Biagini, E.; Blom, N.A.; De Boer, R.A.; et al. 2023 ESC Guidelines for the Management of Cardiomyopathies. Eur. Heart J. 2023, 44, 3503–3626. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, J.P.; Lopes, L.R.; Perez-Barbeito, M.; Cazón-Varela, L.; De La Torre-Carpente, M.M.; Sonicheva-Paterson, N.; De Uña-Iglesias, D.; Quinn, E.; Kuzmina-Krutetskaya, S.; Garrote, J.A.; et al. Deletions of Specific Exons of FHOD3 Detected by Next-generation Sequencing Are Associated with Hypertrophic Cardiomyopathy. Clin. Genet. 2020, 98, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Mates, J.; Mademont-Soler, I.; Fernandez-Falgueras, A.; Sarquella-Brugada, G.; Cesar, S.; Arbelo, E.; García-Álvarez, A.; Jordà, P.; Toro, R.; Coll, M.; et al. Sudden Cardiac Death and Copy Number Variants: What Do We Know after 10 Years of Genetic Analysis? Forensic Sci. Int. Genet. 2020, 47, 102281. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.S.; Ross, S.B.; Skinner, J.R.; Weintraub, R.G.; Ingles, J.; Semsarian, C.; Bagnall, R.D. Characterization of Clinically Relevant Copy-Number Variants from Exomes of Patients with Inherited Heart Disease and Unexplained Sudden Cardiac Death. Genet. Med. 2021, 23, 86–93. [Google Scholar] [CrossRef]
- Lee, C.; Scherer, S.W. The Clinical Context of Copy Number Variation in the Human Genome. Expert Rev. Mol. Med. 2010, 12, e8. [Google Scholar] [CrossRef]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy Number Variation in Human Health, Disease, and Evolution. Annu. Rev. Genom. Hum. Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef] [PubMed]
- Marian, A.J.; Yu, Q.T.; Mares, A.; Hill, R.; Roberts, R.; Perryman, M.B. Detection of a New Mutation in the β-Myosin Heavy Chain Gene in an Individual with Hypertrophic Cardiomyopathy. J. Clin. Investig. 1992, 90, 2156–2165. [Google Scholar] [CrossRef] [PubMed]
- Eddy, C.-A.; MacCormick, J.M.; Chung, S.-K.; Crawford, J.R.; Love, D.R.; Rees, M.I.; Skinner, J.R.; Shelling, A.N. Identification of Large Gene Deletions and Duplications in KCNQ1 and KCNH2 in Patients with Long QT Syndrome. Heart Rhythm 2008, 5, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Mademont-Soler, I.; Mates, J.; Yotti, R.; Espinosa, M.A.; Pérez-Serra, A.; Fernandez-Avila, A.I.; Coll, M.; Méndez, I.; Iglesias, A.; Del Olmo, B.; et al. Additional Value of Screening for Minor Genes and Copy Number Variants in Hypertrophic Cardiomyopathy. PLoS ONE 2017, 12, e0181465. [Google Scholar] [CrossRef]
- Peña-Peña, M.L.; Trujillo-Quintero, J.P.; García-Medina, D.; Cantero-Pérez, E.M.; De Uña-Iglesias, D.; Monserrat, L. Identification by Next-Generation Sequencing of 2 Novel Cases of Noncompaction Cardiomyopathy Associated with 1p36 Deletions. Rev. Española De Cardiol. (Engl. Ed.) 2020, 73, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Rehm, H.L.; Berg, J.S.; Brooks, L.D.; Bustamante, C.D.; Evans, J.P.; Landrum, M.J.; Ledbetter, D.H.; Maglott, D.R.; Martin, C.L.; Nussbaum, R.L.; et al. ClinGen—The Clinical Genome Resource. N. Engl. J. Med. 2015, 372, 2235–2242. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- de Uña-Iglesias, D. System and Method to Detect Structural Genetic Variants [Internet]. Spain: OEPM (Oficina Española de Patentes y Marcas); ES2711163. 2019. Available online: https://consultas2.oepm.es/pdf/ES/0000/000/02/71/11/ES-2711163_B2.pdf (accessed on 9 June 2024).
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical Standards for the Interpretation and Reporting of Constitutional Copy-Number Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Ceyhan-Birsoy, O.; Pugh, T.J.; Bowser, M.J.; Hynes, E.; Frisella, A.L.; Mahanta, L.M.; Lebo, M.S.; Amr, S.S.; Funke, B.H. Next Generation Sequencing-based Copy Number Analysis Reveals Low Prevalence of Deletions and Duplications in 46 Genes Associated with Genetic Cardiomyopathies. Mol. Genet. Genom. Med. 2016, 4, 143–151. [Google Scholar] [CrossRef]
- Lopes, L.R.; Murphy, C.; Syrris, P.; Dalageorgou, C.; McKenna, W.J.; Elliott, P.M.; Plagnol, V. Use of High-Throughput Targeted Exome-Sequencing to Screen for Copy Number Variation in Hypertrophic Cardiomyopathy. Eur. J. Med. Genet. 2015, 58, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Janin, A.; Januel, L.; Cazeneuve, C.; Delinière, A.; Chevalier, P.; Millat, G. Molecular Diagnosis of Inherited Cardiac Diseases in the Era of Next-Generation Sequencing: A Single Center’s Experience Over 5 Years. Mol. Diagn. Ther. 2021, 25, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Alimohamed, M.Z.; Johansson, L.F.; Posafalvi, A.; Boven, L.G.; Van Dijk, K.K.; Walters, L.; Vos, Y.J.; Westers, H.; Hoedemaekers, Y.M.; Sinke, R.J.; et al. Diagnostic Yield of Targeted next Generation Sequencing in 2002 Dutch Cardiomyopathy Patients. Int. J. Cardiol. 2021, 332, 99–104. [Google Scholar] [CrossRef] [PubMed]

| Genes Studied | ||||||
|---|---|---|---|---|---|---|
| AARS2 | ABCC9 | ACAD9 | ACADVL | ACTA1 | ACTC1 | ACTN2 |
| AGK | AGL | AGPAT2 | AKAP9 | ALMS1 | ANK2 | ANK3 |
| ANKRD1 | ATPAF2 | BAG3 | BRAF | BSCL2 | CACNA1C | CACNA1D |
| CACNA2D1 | CACNB2 | CALM1 | CALM2 | CALR3 | CAPN3 | CASQ2 |
| CAV3 | CAVIN4 | COQ2 | COX15 | COX6B1 | CRYAB | CSRP3 |
| CTNNA3 | DES | DLD | DMD | DNAJC19 | DOLK | DSC2 |
| DSG2 | DSP | DTNA | EMD | EYA4 | FAH | FHL1 |
| FHL2 | FHOD3 | FKRP | FKTN | FLNC | FOXD4 | GAA |
| GATA4 | GATA6 | GATAD1 | GFM1 | GJA1 | GJA5 | GLA |
| GLB1 | GNPTAB | GPD1L | GUSB | HCN4 | HFE | HRAS |
| JPH2 | JUP | KCNA5 | KCND3 | KCNE1 | KCNE2 | KCNE3 |
| KCNE5 | KCNH2 | KCNJ2 | KCNJ5 | KCNJ8 | KCNK3 | KCNQ1 |
| KLF10 | KRAS | LAMA2 | LAMA4 | LAMP2 | LDB3 | LDLR |
| LIAS | LMNA | MAP2K1 | MAP2K2 | MIB1 | MLYCD | MRPL3 |
| MRPS22 | MTO1 | MYBPC3 | MYH11 | MYH6 | MYH7 | MYL2 |
| MYL3 | MYLK2 | MYOT | MYOZ2 | MYPN | NEBL | NEXN |
| NKX2-5 | NOTCH1 | NPPA | NRAS | OBSL1 | PDHA1 | PDLIM3 |
| PHKA1 | PITX2 | PKP2 | PLN | PMM2 | PRDM16 | PRKAG2 |
| PSEN1 | PSEN2 | PTPN11 | RAF1 | RANGRF | RBM20 | RYR2 |
| SCN10A | SCN1B | SCN2B | SCN3B | SCN4B | SCN5A | SGCA |
| SGCB | SGCD | SHOC2 | SLC22A5 | SLC25A4 | SLMAP | SNTA1 |
| SOS1 | SPRED1 | SRY | SURF1 | TAZ | TBX20 | TBX5 |
| TCAP | TGFB3 | TMEM43 | TMEM70 | TNNC1 | TNNI3 | TNNT2 |
| TPM1 | TRDN | TRIM63 | TRPM4 | TSFM | TTN | TTR |
| TXNRD2 | VCL | |||||
| Study | CNVs | CM | HCM | DCM | ARVC | LVNC | RCM |
|---|---|---|---|---|---|---|---|
| + | + | 95 | 20 | 50 | 17 | 12 | 1 |
| + | − | 3537 | 2203 | 957 | 279 | 195 | 26 |
| − | − | 8015 | 4576 | 2543 | 606 | 461 | 42 |
| Patients | studied | 11,647 | 6799 | 2550 | 902 | 668 | 69 |
| %CNVs+ | (total) | 0.82 | 0.29 | 1.41 | 1.88 | 1.80 | 1.45 |
| %CNVs+ | (study+) | 2.62 | 0.90 | 4.97 | 5.74 | 5.80 | 3.70 |
| Gene | CM | HCM | DCM | ARVC | LVNC | RCM |
|---|---|---|---|---|---|---|
| DMD | 34 | 31 | 2 | 3 | ||
| PKP2 | 14 | 2 | 1 | 12 | ||
| MYBPC3 | 11 | 11 | 1 | |||
| DSP | 4 | 2 | 1 | 1 | ||
| FLNC | 4 | 2 | 2 | |||
| LAMP2 | 4 | 1 | 3 | |||
| LMNA | 4 | 4 | ||||
| TTN | 4 | 4 | 2 | |||
| FHOD3 | 3 | 3 | ||||
| PLN | 2 | 1 | 1 | |||
| PRDM16 | 2 | 2 | ||||
| RYR2 | 2 | 2 | ||||
| DES | 1 | 1 | ||||
| FHL1 | 1 | 1 | ||||
| MYH7 | 1 | 1 | ||||
| TNX20 | 1 | 1 | ||||
| TBX5 | 1 | 1 | ||||
| TXNRD2 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Uña-Iglesias, D.; Ochoa, J.P.; Monserrat, L.; Barriales-Villa, R. Clinical Relevance of the Systematic Analysis of Copy Number Variants in the Genetic Study of Cardiomyopathies. Genes 2024, 15, 774. https://doi.org/10.3390/genes15060774
de Uña-Iglesias D, Ochoa JP, Monserrat L, Barriales-Villa R. Clinical Relevance of the Systematic Analysis of Copy Number Variants in the Genetic Study of Cardiomyopathies. Genes. 2024; 15(6):774. https://doi.org/10.3390/genes15060774
Chicago/Turabian Stylede Uña-Iglesias, David, Juan Pablo Ochoa, Lorenzo Monserrat, and Roberto Barriales-Villa. 2024. "Clinical Relevance of the Systematic Analysis of Copy Number Variants in the Genetic Study of Cardiomyopathies" Genes 15, no. 6: 774. https://doi.org/10.3390/genes15060774
APA Stylede Uña-Iglesias, D., Ochoa, J. P., Monserrat, L., & Barriales-Villa, R. (2024). Clinical Relevance of the Systematic Analysis of Copy Number Variants in the Genetic Study of Cardiomyopathies. Genes, 15(6), 774. https://doi.org/10.3390/genes15060774






