Placental Transcriptome Analysis in Connection with Low Litter Birth Weight Phenotype (LBWP) Sows
Abstract
1. Introduction
2. Materials and Methods
2.1. Sow Selection and Tissue Collection
2.2. RNA Preparation
2.3. Library Template Construction and Sequencing
2.4. Sequence Mapping and Gene Quantification
2.5. Differentially Expressed Gene Detection
2.6. Functional Enrichment Analysis for DEG and TFBS Prediction
2.7. Quantitative Real-Time RT-PCR Validation
3. Results
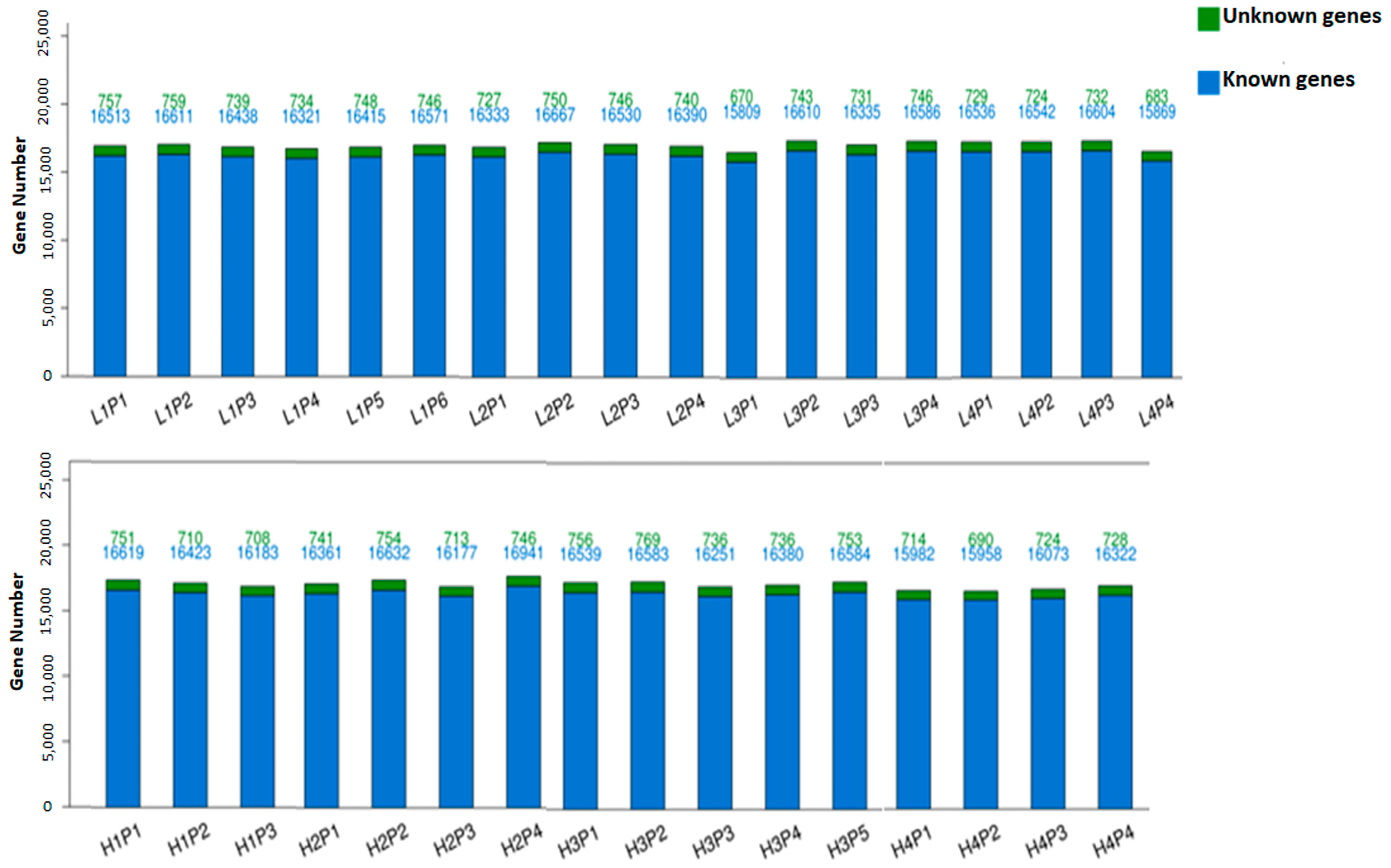
3.1. Sequencing Quality, Transcript Mapping, and Annotation
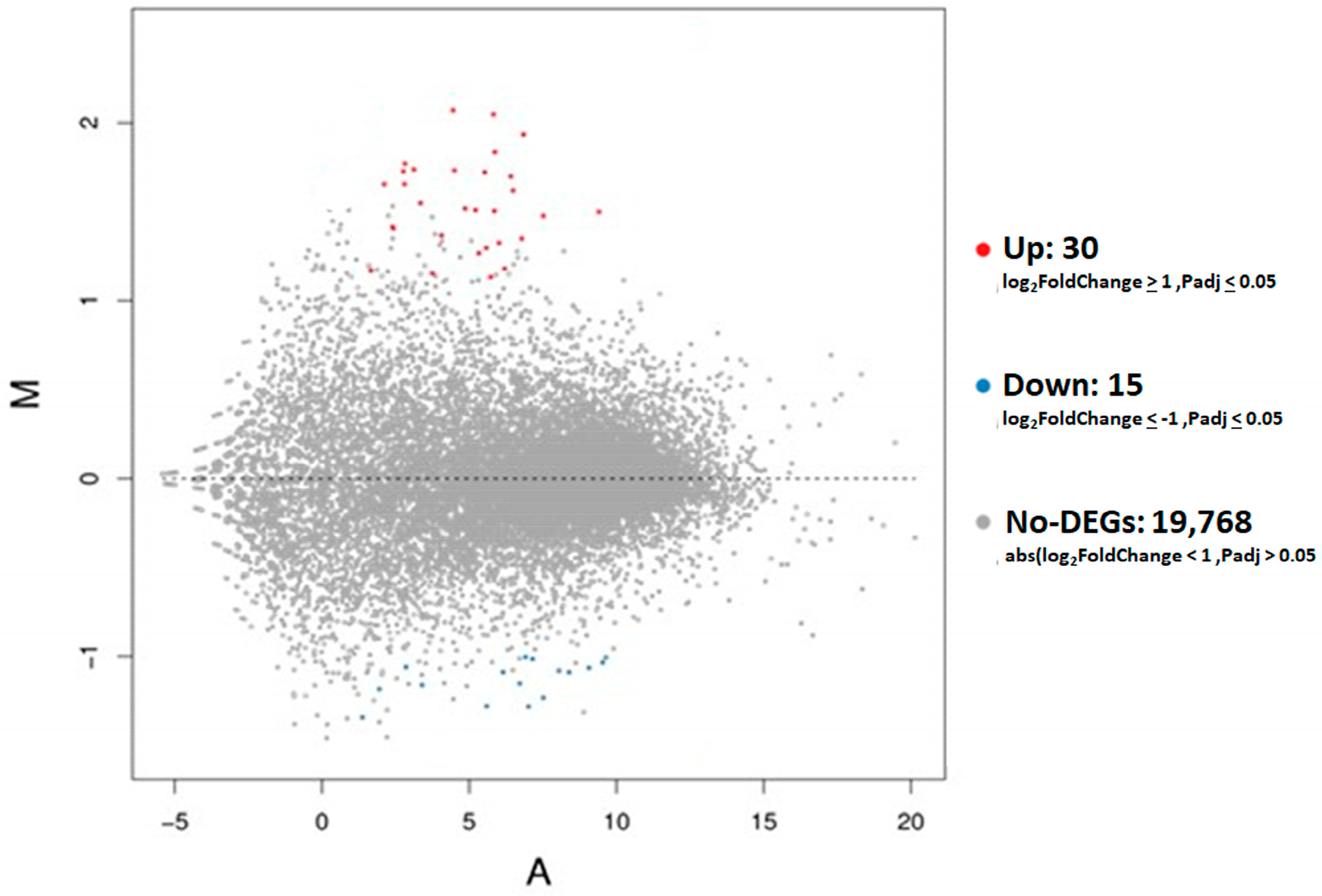
3.2. DEG Analyses in Placental Tissues
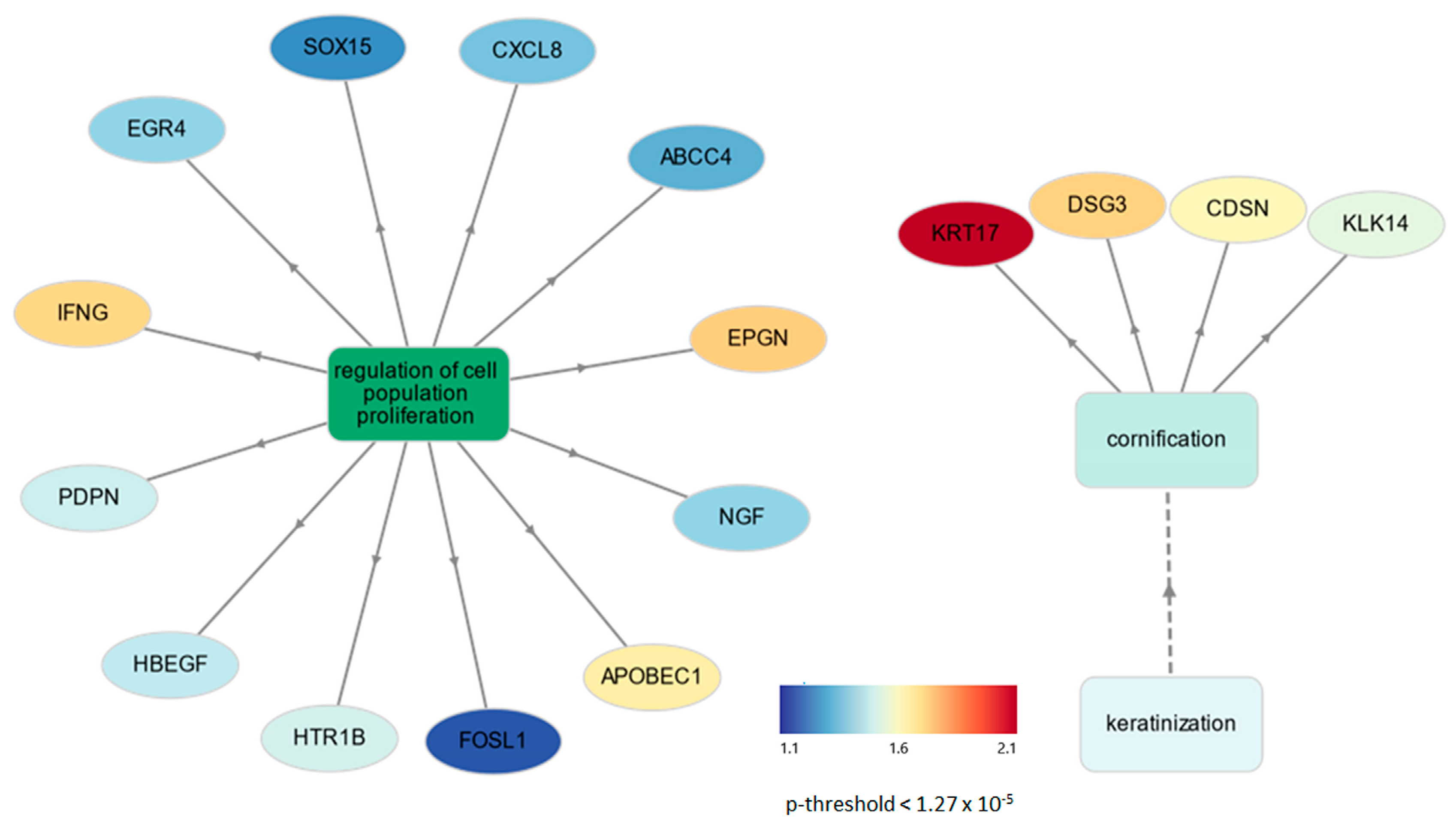
3.3. Functional Enrichment Analysis of DEGs in Placental Tissues
3.4. Transcriptional Factor Binding Site Prediction for Four Genes Involved in Cornification
3.5. Validation of DEG Data by RT-qPCR
4. Discussion
4.1. Genes Differentially Expressed in Placental Tissues from LLBWP Sows
4.2. Impact of Placental Tissue Cornification and Cell Proliferation in LLBWP Sows
4.3. Placental Compensation Mechanisms in LLBWP
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patterson, J.; Foxcroft, G. Gilt Management for Fertility and Longevity. Animals 2019, 9, 434. [Google Scholar] [CrossRef] [PubMed]
- Smit, M.N.; Spencer, J.D.; Almeida, F.R.; Patterson, J.L.; Chiarini-Garcia, H.; Dyck, M.K.; Foxcroft, G.R. Consequences of a low litter birth weight phenotype for postnatal lean growth performance and neonatal testicular morphology in the pig. Animal 2013, 7, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.L.; Smit, M.N.; Novak, S.; Wellen, A.P.; Foxcroft, G.R. Restricted feed intake in lactating primiparous sows. I. Effects on sow metabolic state and subsequent reproductive performance. Reprod. Fertil. Dev. 2011, 23, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Plastow, G.; Charagu, P.; Moroni, J.L.; Patterson, J.; Foxcroft, G.; Dyck, M. Genomic studies of important litter traits in sows. Adv. Pork Prod. 2018, 29, 2767–2778. [Google Scholar]
- Foxcroft, G.R.; Dixon, W.T.; Dyck, M.K.; Novak, S.; Harding, J.C.; Almeida, F.C. Prenatal programming of postnatal development in the pig. Soc. Reprod. Fertil. Suppl. 2009, 66, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, L.P.; Borowicz, P.P.; Caton, J.S.; Vonnahme, K.A.; Luther, J.S.; Buchanan, D.S.; Hafez, S.A.; Grazul-Bilska, A.T.; Redmer, D.A. Uteroplacental vascular development and placental function: An update. Int. J. Dev. Biol. 2010, 54, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Angiolini, E.; Fowden, A.; Coan, P.; Sandovici, I.; Smith, P.; Dean, W.; Burton, G.; Tycko, B.; Reik, W.; Sibley, C.; et al. Regulation of placental efficiency for nutrient transport by imprinted genes. Placenta 2006, 27 (Suppl. A), S98–S102. [Google Scholar] [CrossRef] [PubMed]
- Jessmon, P.; Leach, R.E.; Armant, D.R. Diverse functions of HBEGF during pregnancy. Mol. Reprod. Dev. 2009, 76, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Moroni, J.L.; Tsoi, S.; Wenger, I.; Tran, C.; Plastow, G.S.; Charagu, P.; Dyck, M.K. The influence of litter birth weight phenotype on embryonic and placental development at day 30 of gestation in multiparous purebred Large White sows. Anim. Reprod. Sci. 2022, 244, 107035. [Google Scholar] [CrossRef]
- Snelling, W.M.; Cushman, R.A.; Keele, J.W.; Maltecca, C.; Thomas, M.G.; Fortes, M.R.; Reverter, A. Breeding and Genetics Symposium: Networks and pathways to guide genomic selection. J. Anim. Sci. 2013, 91, 537–552. [Google Scholar] [CrossRef]
- Karisa, B.; Moore, S.; Plastow, G. Analysis of biological networks and biological pathways associated with residual feed intake in beef cattle. Anim. Sci. J. 2014, 85, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Steibel, J.P.; Bates, R.O.; Rosa, G.J.; Tempelman, R.J.; Rilington, V.D.; Ragavendran, A.; Raney, N.E.; Ramos, A.M.; Cardoso, F.F.; Edwards, D.B.; et al. Genome-wide linkage analysis of global gene expression in loin muscle tissue identifies candidate genes in pigs. PLoS ONE 2011, 6, e16766. [Google Scholar] [CrossRef] [PubMed]
- Spotter, A.; Distl, O. Genetic approaches to the improvement of fertility traits in the pig. Vet. J. 2006, 172, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, X.; Michal, J.J.; Ding, B.; Li, R.; Jiang, Z. Genome wide screening of candidate genes for improving piglet birth weight using high and low estimated breeding value populations. Int. J. Biol. Sci. 2014, 10, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Blanes, M.S.; Tsoi, S.C.; Dyck, M.K. Accurate and Phenol Free DNA Sexing of Day 30 Porcine Embryos by PCR. J. Vis. Exp. 2016, 108, e53301. [Google Scholar] [CrossRef] [PubMed]
- Fehlmann, T.; Reinheimer, S.; Geng, C.; Su, X.; Drmanac, S.; Alexeev, A.; Zhang, C.; Backes, C.; Ludwig, N.; Hart, M.; et al. cPAS-based sequencing on the BGISEQ-500 to explore small non-coding RNAs. Clin. Epigenetics 2016, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Pomaznoy, M.; Ha, B.; Peters, B. GOnet: A tool for interactive Gene Ontology analysis. BMC Bioinform. 2018, 19, 470. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hu, H.; Miao, Y.R.; Jia, L.H.; Yu, Q.Y.; Zhang, Q.; Guo, A.Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Pavlicev, M.; Wagner, G.P.; Chavan, A.R.; Owens, K.; Maziarz, J.; Dunn-Fletcher, C.; Kallapur, S.G.; Muglia, L.; Jones, H. Single-cell transcriptomics of the human placenta: Inferring the cell communication network of the maternal-fetal interface. Genome Res. 2017, 27, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.C.; Miles, J.R.; Lents, C.A.; Rempel, L.A. Uterine and placenta characteristics during early vascular development in the pig from day 22 to 42 of gestation. Anim. Reprod. Sci. 2016, 164, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Tayade, C.; Fang, Y.; Croy, B.A. A review of gene expression in porcine endometrial lymphocytes, endothelium and trophoblast during pregnancy success and failure. J. Reprod. Dev. 2007, 53, 455–463. [Google Scholar] [CrossRef]
- Stenhouse, C.; Hogg, C.O.; Ashworth, C.J. Associations between fetal size, sex and placental angiogenesis in the pig. Biol. Reprod. 2019, 100, 239–252. [Google Scholar] [CrossRef]
- Stenhouse, C.; Hogg, C.O.; Ashworth, C.J. Novel relationships between porcine fetal size, sex, and endometrial angiogenesisdagger. Biol. Reprod. 2019, 101, 112–125. [Google Scholar] [CrossRef]
- Freitag, L.; von Kaisenberg, C.; Kreipe, H.; Hussein, K. Expression analysis of leukocytes attracting cytokines in chronic histiocytic intervillositis of the placenta. Int. J. Clin. Exp. Pathol. 2013, 6, 1103–1111. [Google Scholar] [PubMed]
- Onak Kandemir, N.; Barut, F.; Barut, A.; Birol, I.E.; Dogan Gun, B.; Ozdamar, S.O. Biological importance of podoplanin expression in chorionic villous stromal cells and its relationship to placental pathologies. Sci. Rep. 2019, 9, 14230. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, J.; Gu, Y.; Zhao, S.; Groome, L.J.; Alexander, J.S. D2-40/podoplanin expression in the human placenta. Placenta 2011, 32, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Ishida-Yamamoto, A.; Igawa, S. The biology and regulation of corneodesmosomes. Cell Tissue Res. 2015, 360, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Gomez, T.; Ona, K.; Kapidzic, M.; Gormley, M.; Simon, C.; Genbacev, O.; Fisher, S.J. Severe pre-eclampsia is associated with alterations in cytotrophoblasts of the smooth chorion. Development 2017, 144, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Eckhart, L.; Lippens, S.; Tschachler, E.; Declercq, W. Cell death by cornification. Biochim. Biophys. Acta 2013, 1833, 3471–3480. [Google Scholar] [CrossRef] [PubMed]
- Mills, I.H. Kallikrein, kininogen and kinins in control of blood pressure. Nephron 1979, 23, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.D.; Molina, L.; Bhoola, K.D.; Ehrenfeld, P. Overview of tissue kallikrein and kallikrein-related peptidases in breast cancer. Biol. Chem. 2018, 399, 937–957. [Google Scholar] [CrossRef]
- Fernando, S.C.; Buck, J.S.; Ashworth, M.D.; Ross, J.W.; Geisert, R.D.; DeSilva, U. Porcine endometrial and conceptus tissue kallikrein 1, 4, 11, and 14 gene expression. Reproduction 2006, 132, 939–947. [Google Scholar] [CrossRef]
- Depianto, D.; Kerns, M.L.; Dlugosz, A.A.; Coulombe, P.A. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat. Genet. 2010, 42, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Charo, I.F.; Ransohoff, R.M. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 2006, 354, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Drake, P.M.; Red-Horse, K.; Fisher, S.J. Chemokine expression and function at the human maternal-fetal interface. Rev. Endocr. Metab. Disord. 2002, 3, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Hannan, N.J.; Salamonsen, L.A. Role of chemokines in the endometrium and in embryo implantation. Curr. Opin. Obstet. Gynecol. 2007, 19, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Wang, S.; Li, D. The integrative roles of chemokines at the maternal–fetal interface in early pregnancy. Cell. Mol. Immunol. 2014, 11, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Warning, J.C.; McCracken, S.A.; Morris, J.M. A balancing act: Mechanisms by which the fetus avoids rejection by the maternal immune system. Reproduction 2011, 141, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Polgar, K.; Hill, J.A. Identification of the white blood cell populations responsible for Th1 immunity to trophoblast and the timing of the response in women with recurrent pregnancy loss. Gynecol. Obstet. Investig. 2002, 53, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Dent, L.A. For better or worse: Common determinants influencing health and disease in parasitic infections, asthma and reproductive biology. J. Reprod. Immunol. 2002, 57, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Patrick, L.A.; Smith, G.N. Proinflammatory cytokines: A link between chorioamnionitis and fetal brain injury. J. Obstet. Gynaecol. Can. 2002, 24, 705–709. [Google Scholar] [CrossRef]
- Stemmer, S.M. Current clinical progress in early pregnancy investigation. Early Pregnancy 2000, 4, 74–81. [Google Scholar]
- Croy, B.A.; Wessels, J.; Linton, N.; Tayade, C. Comparison of immune cell recruitment and function in endometrium during development of epitheliochorial (pig) and hemochorial (mouse and human) placentas. Placenta 2009, 30 (Suppl. A), S26–S31. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert. Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Sakai, M.; Sasaki, Y.; Yoneda, S.; Kasahara, T.; Arai, T.; Okada, M.; Hosokawa, H.; Kato, K.; Soeda, Y.; Saito, S. Elevated interleukin-8 in cervical mucus as an indicator for treatment to prevent premature birth and preterm, pre-labor rupture of membranes: A prospective study. Am. J. Reprod. Immunol. 2004, 51, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Szarka, A.; Rigo, J., Jr.; Lazar, L.; Beko, G.; Molvarec, A. Circulating cytokines, chemokines and adhesion molecules in normal pregnancy and preeclampsia determined by multiplex suspension array. BMC Immunol. 2010, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.P.; Tayade, C.; Ashkar, A.A.; Hatta, K.; Zhang, J.; Croy, B.A. Interferon gamma in successful pregnancies. Biol. Reprod. 2009, 80, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Jana, B.; Andronowska, A. Expression of nerve growth factor and its receptors in the uterus of gilts with endometritis induced by infection with Escherichia coli. J. Comp. Pathol. 2012, 147, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Vallet, J.L.; Freking, B.A. Differences in placental structure during gestation associated with large and small pig fetuses. J. Anim. Sci. 2007, 85, 3267–3275. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Johnson, G.A. Pig blastocyst-uterine interactions. Differentiation 2014, 87, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, T.G.; Brown, K.D.; Vaughan, T.J. Expression of the genes for the epidermal growth factor receptor and its ligands in porcine oviduct and endometrium. Biol. Reprod. 1994, 50, 751–756. [Google Scholar] [CrossRef]
- Iwamoto, R.; Yamazaki, S.; Asakura, M.; Takashima, S.; Hasuwa, H.; Miyado, K.; Adachi, S.; Kitakaze, M.; Hashimoto, K.; Raab, G.; et al. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc. Natl. Acad. Sci. USA 2003, 100, 3221–3226. [Google Scholar] [CrossRef]
- Vonnahme, K.A.; Wilson, M.E.; Ford, S.P. Relationship between placental vascular endothelial growth factor expression and placental/endometrial vascularity in the pig. Biol. Reprod. 2001, 64, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, L.A.; Schreier, L.L.; Guthrie, H.D.; Sample, G.L.; Vallet, J.; Caperna, T.; Ramsay, T. The effect of intrauterine growth retardation on the expression of developmental factors in porcine placenta subsequent to the initiation of placentation. Placenta 2010, 31, 549–552. [Google Scholar] [CrossRef] [PubMed]
| RNAseq | RT-qPCR | |||
|---|---|---|---|---|
| Genes | log2FoldChange | p-Value | log2FoldChange | p-Value |
| (LLBWP/HLBWP) | (LLBWP/HLBWP) | |||
| CDSN | 1.62 | 0.0165551 | 3.55 | 0.0057 |
| HBEGF | 1.48 | 0.0110607 | 3.33 | 0.002 |
| PDPN | 1.5 | 0.003 | 2.14 | 0.0018 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linck Moroni, J.; Tsoi, S.; Wenger, I.I.; Plastow, G.S.; Dyck, M.K. Placental Transcriptome Analysis in Connection with Low Litter Birth Weight Phenotype (LBWP) Sows. Genes 2024, 15, 703. https://doi.org/10.3390/genes15060703
Linck Moroni J, Tsoi S, Wenger II, Plastow GS, Dyck MK. Placental Transcriptome Analysis in Connection with Low Litter Birth Weight Phenotype (LBWP) Sows. Genes. 2024; 15(6):703. https://doi.org/10.3390/genes15060703
Chicago/Turabian StyleLinck Moroni, Julia, Stephen Tsoi, Irene I. Wenger, Graham S. Plastow, and Michael K. Dyck. 2024. "Placental Transcriptome Analysis in Connection with Low Litter Birth Weight Phenotype (LBWP) Sows" Genes 15, no. 6: 703. https://doi.org/10.3390/genes15060703
APA StyleLinck Moroni, J., Tsoi, S., Wenger, I. I., Plastow, G. S., & Dyck, M. K. (2024). Placental Transcriptome Analysis in Connection with Low Litter Birth Weight Phenotype (LBWP) Sows. Genes, 15(6), 703. https://doi.org/10.3390/genes15060703







