Exploring the Role of the MUTYH Gene in Breast, Ovarian and Endometrial Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Germline DNA Testing
2.2. Somatic DNA Testing
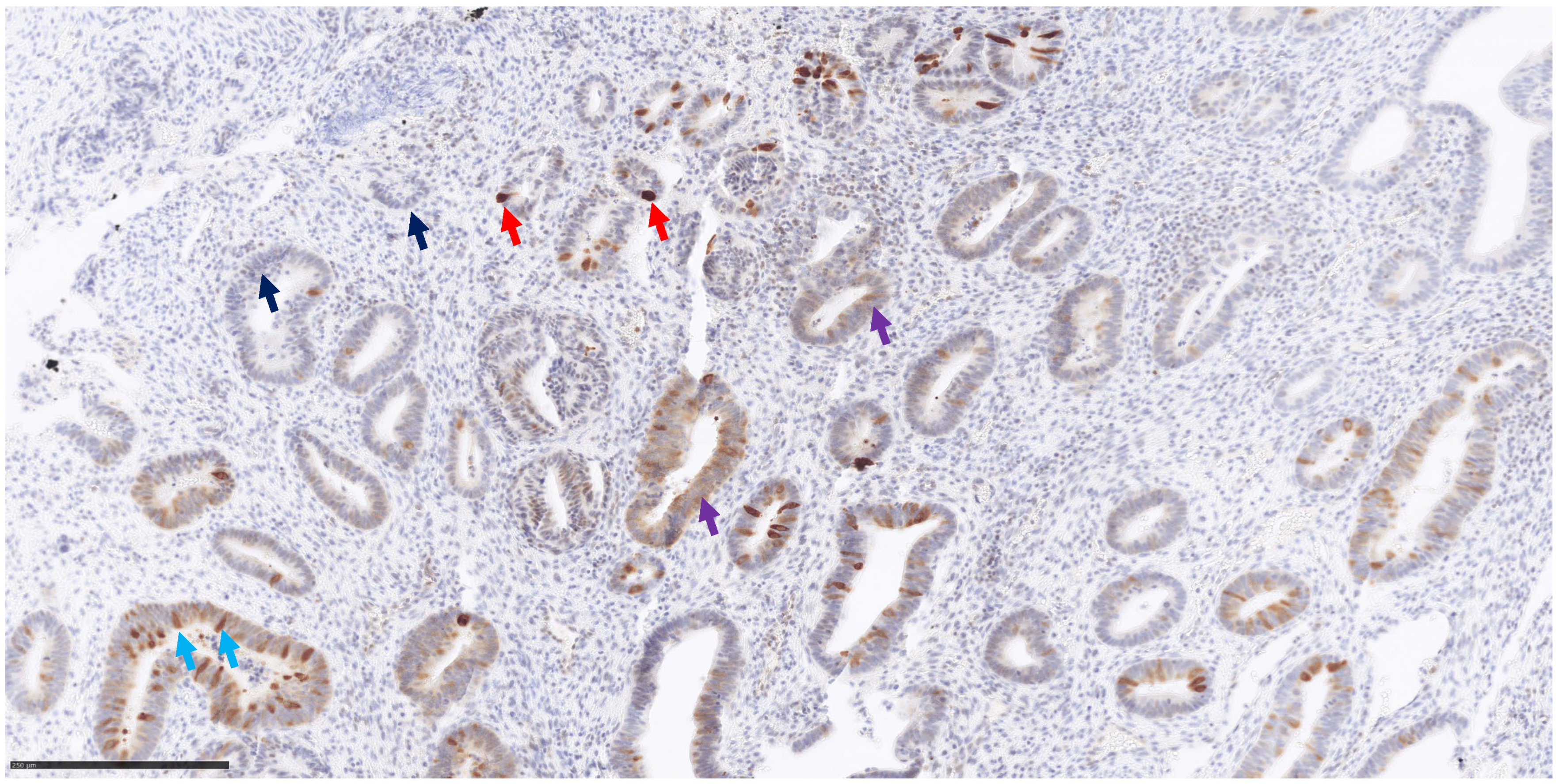
2.3. MUTYH Immunohistochemistry
3. Results
3.1. MUTYH Germline Variants
3.2. MUTYH Somatic Variants
3.3. MUTHY Immunohistochemistry
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Disclai Cheadle, J.P.; Sampson, J.R. Exposing the MYtH about base excision repair and human inherited disease. Hum. Molec. Genet. 2003, 12, R159–R165. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.R.; Eshleman, J.R. Human MutY: Gene structure, protein functions and interactions, and role in carcinogenesis. Cell. Mol. Life Sci. 2003, 60, 2064–2083. [Google Scholar] [CrossRef] [PubMed]
- Babaei, K.; Khaksar, R.; Zeinali, T.; Hemmati, H.; Bandegi, A.; Samidoust, P.; Ashoobi, M.T.; Hashemian, H.; Delpasand, K.; Talebinasab, F.; et al. Epigenetic profiling of MUTYH, KLF6, WNT1 and KLF4 genes in carcinogenesis and tumorigenesis of colorectal cancer. Biomedicine 2019, 9, 22. [Google Scholar] [CrossRef]
- Nunziato, M.; Di Maggio, F.; Pensabene, M.; Esposito, M.V.; Starnone, F.; De Angelis, C.; Calabrese, A.; D’Aiuto, M.; Botti, G.; De Placido, S.; et al. Multi-gene panel testing increases germline predisposing mutations’ detection in a cohort of breast/ovarian cancer patients from Southern Italy. Front. Med. 2022, 9, 894358. [Google Scholar] [CrossRef] [PubMed]
- Chávarri-Guerra, Y.; Bourlon, M.T.; Rodríguez-Olivares, J.L.; Orozco, L.; Bazua, D.; Rodríguez-Faure, A.; Alcalde-Castro, M.J.; Castro, E.; Castillo, D.; Herzog, J.; et al. Germline DNA Repair Genes Pathogenic Variants Among Mexican Patients with Prostate Cancer. Clin. Genitourin. Cancer 2023, 21, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Lintas, C.; Facchiano, A.; Azzarà, A.; Cassano, I.; Tabolacci, C.; Galasso, C.; Gurrieri, F. Deletion of a Single Lysine Residue at Position 292 of CAMK2A Disrupts Protein Function, Causing Severe Epileptic Encephalopathy and Intellectual Disability. Genes 2023, 14, 1353. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Tadrous, P.J. Breast. In Diagnostic Criteria Handbook in Histopathology: A Surgical Pathology Vade Mecum; John Wiley & Sons: Oxford, UK, 2008; pp. 258–266. [Google Scholar]
- Mak, S.; Alexander, J.L.; Clark, S.K.; Hawkins, M.; Cuthill, V.; Latchford, A.; Monahan, K.J. The Diagnostic Yield of Genetic Testing in Patients with Multiple Colorectal Adenomas: A Specialist Center Cohort Study. Clin. Transl. Gastroenterol. 2024, 15, e00645. [Google Scholar] [CrossRef] [PubMed]
- Di Gregorio, C.; Frattini, M.; Maffei, S.; Ponti, G.; Losi, L.; Pedroni, M.; Venesio, T.; Bertario, L.; Varesco, L.; Risio, M.; et al. Immunohistochemical expression of MYH protein can be used to identify patients with MYH-associated polyposis. Gastroenterology 2006, 131, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Van der Post, R.S.; Kets, C.M.; Ligtenberg, M.J.; van Krieken, J.H.; Hoogerbrugge, N. Immunohistochemistry is not an accurate first step towards the molecular diagnosis of MUTYH-associated polyposis. Virchows Arch. 2009, 454, 25–29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oliveira, P.; Correa, P.; Acosta, A.; Freitas, J.; Machado-Lopes, T.; Bomfim-Palma, T.; Ribeiro-Dos-Santos, Â.; Santos, S.; Nascimento, R.; Nascimento, I.; et al. Screening for Mutations in Hereditary Cancer Susceptibility Genes in a Region with High Endogamy in Brazil. Glob. Med. Genet. 2023, 10, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Siegelmann-Danieli, N.; Neiman, V.; Bareket-Samish, A.; Berger, R.; Peretz, A.; Alapi, H.; Tsur, E.; Patalon, T.; Beller, D.; Rimler, G.; et al. Whole exome germline sequencing in early-onset prostate cancer patients: Genomic findings and clinical outcomes. Prostate 2024, 84, 39–46. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.F.; Prather, L.L.; Helfer, C.R.; Ludmir, E.B.; Echeverria, A.E.; Yust-Katz, S.; Patel, A.J.; Deneen, B.; Rao, G.; Jalali, A.; et al. Prevalence of pathogenic germline variants in adult-type diffuse glioma. Neurooncol. Pract. 2023, 10, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Keske, A.; Weisman, P.; Ospina-Romero, M.; Raut, P.; Smith-Simmer, K.; Zakas, A.L.; Flynn, C.; Xu, J. Breast cancers in monoallelic MUTYH germline mutation carriers have clinicopathological features overlapping with those in BRCA1 germline mutation carriers. Breast Cancer Res. Treat. 2024, 204, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, M.A.; Croitoru, M.E.; Monga, N.; Cleary, S.P.; Cotterchio, M.; Hopper, J.L.; Gallinger, S. Risk of colorectal cancer in monoallelic and biallelic carriers of MYH mutations: A population-based case-family study. Cancer Epidemiol. Biomark. Prev. 2006, 15, 312–314. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.; Vogt, S.; Nielsen, M.; Christian, D.; Wark, P.A.; Eccles, D.; Edwards, E.; Evans, D.G.; Maher, E.R.; Vasen, H.F.; et al. Increased colorectal cancer incidence in obligate carriers of heterozygous mutations in MUTYH. Gastroenterology 2009, 137, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.B.; Sutcliffe, E.G.; Arvai, K.; Roberts, M.E.; Susswein, L.R.; Marshall, M.L.; Torene, R.; Postula, K.J.V.; Hruska, K.S.; Bai, S. Monoallelic MUTYH pathogenic variants ascertained via multi-gene hereditary cancer panels are not associated with colorectal, endometrial, or breast cancer. Fam. Cancer 2022, 21, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, M.L.; Zhao, E.Y.; Reisle, C.; Ch’ng, C.; Wong, H.L.; Shen, Y.; Jones, M.R.; Lim, H.J.; Young, S.; Cremin, C.; et al. Base excision repair deficiency signatures implicate germline and somatic MUTYH aberrations in pancreatic ductal adenocarcinoma and breast cancer oncogenesis. Cold Spring Harb. Mol. Case Stud. 2019, 5, a003681. [Google Scholar] [CrossRef] [PubMed]
- Nones, K.; Johnson, J.; Newell, F.; Patch, A.M.; Thorne, H.; Kazakoff, S.H.; de Luca, X.M.; Parsons, M.T.; Ferguson, K.; Reid, L.E.; et al. Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann. Oncol. 2019, 30, 1071–1079. [Google Scholar] [CrossRef]

| ID | Birth Date | Age at Cancer Diagnosis | Familiarity for Neoplasms * | % of Neoplastic Cells in the Tumor Sample | Diagnosis |
|---|---|---|---|---|---|
| pz 1 | 3 July 1943 | 76 | positive | 60 | breast and other tumors |
| pz 2 | 30 June 1948 | 72 | positive | 30 | ovarian carcinoma |
| pz 3 | 15 October 1979 | 37 | positive | 60 | adenocarcinoma of endometrium extended to ovary |
| pz 4 | 6 January 1979 | 41 | positive | 30 | breast carcinoma |
| pz 5 | 11 January 1970 | 44 | positive | 20 | breast carcinoma |
| pz 6 | 24 March 1961 | 60 | negative | 80 | adenocarcinoma of endometrium |
| pz 7 | 23 November 1975 | 42 | positive | 60 | bilateral breast carcinoma |
| pz 8 | 8 December 1997 | 23 | positive | 40 | bilateral ovarian carcinoma |
| pz 9 | 4 August 1966 | 55 | positive | 40 | adenocarcinoma of endometrium |
| Panel A | ||||||
| ID | Germinal Variant | Exon | ClinVar Class | AMCG Class | Variants in Other Genes | |
| pz 1 | c.1187 G>A p.Gly396Asp | 13 | LP/P | P | / | |
| pz 2 | c.1187 G>A p.Gly396Asp | 13 | LP/P | P | BRCA1 VUS (c.5332 G>A, p. Asp1178Asn) | |
| pz 3 | c. 536A>G p.Tyr179Cys | 7 | LP/P | P | / | |
| pz 4 | c.1187 G>A p.Gly396Asp | 13 | LP/P | P | / | |
| pz 5 | c. 536A>G p.Tyr179Cys | 7 | LP/P | P | BRCA2 VUS (c.3680 T>C p.Leu1227Pro) and CDH1 VUS (c.74C>T p. Pro25Leu) | |
| pz 6 | c. 536A>G p.Tyr179Cys | 7 | LP/P | P | / | |
| pz 7 | c.1187 G>A p.Gly396Asp | 13 | LP/P | P | PALB2 VUS (c.398 G>A p.Ser133Asn) and ATM VUS (c.1516 G>T p.Gly506Cys) | |
| pz 8 | c.1258 C>A p.Leu420Met | 13 | CIP | VUS | / | |
| pz 8 | c.694 A>T p.Thr232Ser (in cis with the other variant) | 9 | VUS | VUS | / | |
| pz 9 | c.1437_1439del p.Glu480del | 14 | P | P | POLE VUS (c.6445 C>T p.Arg2149Cys) | |
| Panel B | ||||||
| ID | Variant | Exon | GnomAD Freq | ClinVar | ACMG Classification | In Silico Predictors |
| pz 1 | / | / | / | / | / | / |
| pz 2 | / | / | / | / | / | / |
| pz 3 | c.770 T>C p.Leu257Pro | 9 | absent | / | VUS | 4VUS/35B |
| pz 3 | c.1256 C>T p. Ala419Val | 13 | 0.00003184 | VUS | VUS | 1P/6VUS/17B |
| pz 4 | / | / | / | / | / | / |
| pz 5 | c.312 C>T p.Tyr104Tyr | 3 | 0.001488 | CIP (1VUS/18LB/7B) | LB | 2B |
| pz 6 | / | / | / | / | / | / |
| pz 7 | c.420 C>T p.Thr140Thr | 5 | absent | LB | LB | 2B |
| pz 8 | c.1014 G>C p.Glu338His | 12 | 0.286 | B | B | 1VUS/29B |
| pz 9 | c.1014 G>C p.Glu338His | 12 | 0.286 | B | B | 1VUS/29B |
| ID | SCORE 0 | SCORE 1+ | SCORE 2+ | SCORE 3+ | IHC Score |
|---|---|---|---|---|---|
| pz 1 | 60 | 20 | 10 | 10 | 70 |
| pz 2 | 100 | 0 | 0 | 0 | 0 |
| pz 3 | 50 | 30 | 10 | 10 | 80 |
| pz 4 | 100 | 0 | 0 | 0 | 0 |
| pz 5 | 100 | 0 | 0 | 0 | 0 |
| pz 6 | 70 | 5 | 5 | 20 | 75 |
| pz 7 | 20 | 5 | 15 | 60 | 215 |
| pz 8 | 100 | 0 | 0 | 0 | 0 |
| pz 9 | 60 | 10 | 10 | 20 | 90 |
| control 1 | 100 | 0 | 0 | 0 | 0 |
| control 2 | 100 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lintas, C.; Canalis, B.; Azzarà, A.; Sabarese, G.; Perrone, G.; Gurrieri, F. Exploring the Role of the MUTYH Gene in Breast, Ovarian and Endometrial Cancer. Genes 2024, 15, 554. https://doi.org/10.3390/genes15050554
Lintas C, Canalis B, Azzarà A, Sabarese G, Perrone G, Gurrieri F. Exploring the Role of the MUTYH Gene in Breast, Ovarian and Endometrial Cancer. Genes. 2024; 15(5):554. https://doi.org/10.3390/genes15050554
Chicago/Turabian StyleLintas, Carla, Benedetta Canalis, Alessia Azzarà, Giovanna Sabarese, Giuseppe Perrone, and Fiorella Gurrieri. 2024. "Exploring the Role of the MUTYH Gene in Breast, Ovarian and Endometrial Cancer" Genes 15, no. 5: 554. https://doi.org/10.3390/genes15050554
APA StyleLintas, C., Canalis, B., Azzarà, A., Sabarese, G., Perrone, G., & Gurrieri, F. (2024). Exploring the Role of the MUTYH Gene in Breast, Ovarian and Endometrial Cancer. Genes, 15(5), 554. https://doi.org/10.3390/genes15050554








