Abstract
Kohlrabi is an important swollen-stem cabbage variety belonging to the Brassicaceae family. However, few complete chloroplast genome sequences of this genus have been reported. Here, a complete chloroplast genome with a quadripartite cycle of 153,364 bp was obtained. A total of 132 genes were identified, including 87 protein-coding genes, 37 transfer RNA genes and eight ribosomal RNA genes. The base composition analysis showed that the overall GC content was 36.36% of the complete chloroplast genome sequence. Relative synonymous codon usage frequency (RSCU) analysis showed that most codons with values greater than 1 ended with A or U, while most codons with values less than 1 ended with C or G. Thirty-five scattered repeats were identified and most of them were distributed in the large single-copy (LSC) region. A total of 290 simple sequence repeats (SSRs) were found and 188 of them were distributed in the LSC region. Phylogenetic relationship analysis showed that five Brassica oleracea subspecies were clustered into one group and the kohlrabi chloroplast genome was closely related to that of B. oleracea var. botrytis. Our results provide a basis for understanding chloroplast-dependent metabolic studies and provide new insight for understanding the polyploidization of Brassicaceae species.
1. Introduction
Kohlrabi (Brassica oleracea var. gongylodes Linnaeus, 1753), an important swollen-stem vegetable variety of B. oleracea variety, originated from northwestern Europe and is widely cultivated in Europe, the US, Canada, and Asia [1]. The swollen stem at the base of the plant is mainly consumed by humans as food [2,3]. Studies have shown that the swollen stem of kohlrabi has high nutritional value, particularly in vitamin C, vitamin E, and tocopherols [4,5,6]. In addition, potential antidiabetic, anti-inflammatory, and antioxidant properties and anticancer effects have been found in kohlrabi [7].
Chloroplasts (cp) are centers of plenty cellular reactions and crucial organelles of plant cells [8,9], originating from photosynthetic cyanobacteria engulfed and enslaved by eukaryotic cells [10,11]. Chloroplasts not only play vital roles in photosynthesis but also contain all the elements required for carbohydrate metabolism and biosynthesis of nucleotides and amino acids [12]. In addition, chloroplasts are involved in various molecular processes, such as regulation of plant physiology, growth, development, and stress responses [12,13,14].
The cp genome is a maternally inherited genetic systems of plants, which is not affected by karyogene deletion, overlap, or pseudogenes [15]. In the cp genome, a large number of mutational events also occur, such as nucleotide substitutions, insertions, deletions, and genome fragment inversions, translocations and rearrangements [16,17,18]. In most angiosperms, chloroplasts have a typical quadripartite circular genome, which comprises one large single-copy (LSC) region with a length of about 81–90 kb, one small single-copy (SSC) region of approximately 18–20 kb, and two inverted repeat (IR) regions of about 20–30 kb, named IRa and IRa [13,15,19]. Oldenburg and Bendich [20] reported that a linear cp genome was found in maize. As plant-specific organelles, chloroplasts have highly conserved genomes in terms of gene content and organization while maintaining a relatively simple structure, small molecular weight and large copy number [21,22,23], owing to a high conservation of structure, moderate genome size and good collinearity between various plant groups of cp genomes [24], which have been widely used in DNA fingerprint development, phylogenetic discrepancy analysis, molecular evolution, and genetic engineering modification, such as B. rapa. ssp. rapa [25], Paeonia ostii [26], Zingiber officinale [27], Withania somnifera [28], Adrinandra megaphylla Hu [29], and Cornus species [30]. Moreover, cp genomes have also been used to deal with important scientific issues of crop origin and domestication [31].
Several cp genome sequence have been reported in B. oleracea: B. oleracea var. botrytis (cauliflower) (KX681655.1), B. oleracea var. italica (cabbage) (MH388765, MN649876.1), B. oleracea var. capitata (red cabbage) (KR233156.1), B. oleracea (wild cabbage) (MG717288) and B. oleracea var. alboglabra (white-flower Chinese kale and yellow-flower Chinese kale) (OR063915 and OR063916). However, despite being the only swollen-stem vegetable of the B. oleracea variety, no sequence of the kohlrabi cp genome has been presented to our knowledge. Thus, the complete sequence of the kohlrabi cp genome was obtained and analyzed in this study.
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
Kohlrabi seeds were sown in holed plates in late April and cultured in greenhouse for 35 days until the seedlings had 4 or 5 leaves, then the seedlings were transplanted in a randomized field plot with regular management in an experimental field (36°420 N; 101°450 E) of the Academy of Agriculture and Forestry Sciences, Qinghai University. The fresh young kohlrabi leaves were collected and the modified CTAB (cetyl trimethyl ammonium bromide) method as Porebski et al. [32] described was used for total DNA extraction. The same DNA samples of Shao et al. [33] were used in this study and stored at the Qinghai Key Laboratory of Vegetable Genetics and Physiology. The kohlrabi plant were showed in Figure 1 and used in this paper.

Figure 1.
The kohlrabi plant grown in the field and used in this paper.
2.2. DNA Sequencing and Assembly
The extracted DNA were assigned for purification and assessment, and then qualifying DNA were used to build a library. This library was sequenced using the Illumina NovaSeq2500 platform (Shaanxi Breeding Biotechnologies Co., Ltd., Xi’an, China). Using SPAdes v3.10.1 [34] software, the cp genome sequence was assembled and did not depend on the reference genome. Then, gap repair in the scaffolds was performed using GapFiller v2.1.1 [35] until a complete pseudogenome was obtained. Finally, based on the structure of chloroplasts and rearrangement of the pseudo genome, a complete cp circular genome was obtained.
2.3. Gene Annotation
Based on the assembled sequences of relative species uploaded to the National Center for Biotechnology Information (NCBI), the online software BLAST v2.6 (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 23 March 2023) was used for gene alignment. Two strategies were used to annotate the cp genome for improving annotation accuracy. Firstly, the prodigal v2.6.3 (https://www.github.com/hyattpd/Prodigal, accessed on 23 March 2023), hmmer v3.1b2 (http://www.hmmer.org/, accessed on 23 March 2023) and aragorn v1.2.38 (http://130.235.244.92/ARAGORN/, accessed on 23 March 2023) were separately used to annotate the coding sequence (CDS), predict the ribosomal RNA (rRNA) and transfer RNA (tRNA). Secondly, based on the cp genomes of closely related species published on NCBI, we obtained annotation information by comparing the assembled sequences using BLAST v2.6 DOGMA (http://dogma.ccbb.utexas.edu/, accessed on 23 March 2023) [36]. Results from two strategies were manually checked for differentially annotated genes. Then, we removed any misannotation and redundant annotation and determined the multi-exon boundaries. Finally, the final annotation information was obtained. The tRNAs were analyzed based on the online software tool tRNAscan-SE with default settings (http://lowelab.ucsc.edu/tRNAscan-SE/, accessed on 23 March 2023) [37]. OrganellarGenomeDRAW (http://ogdraw.mpimp-golm.mpg.de/index.shtml, accessed on 23 March 2023) was applied for visualization of the complete cp genome map [38].
2.4. Codon Usage Frequency Analysis
According to the degeneracy of codons, each amino acid was coded by 1 codon at least and 6 codons at most. In different species and different organisms, there are great differences in genome codon usage rates. The inequality of synonymous codon usage is called relative synonymous codon usage (RESU). RESU was calculated using the following formula: RESU = ratio of (number of one of the codons encoding a certain amino acid) to (number of all codons encoding this amino acid)/(1/the codon species encoding this amino acid).
2.5. Repeat Sequence Analysis
The interspersed repeat sequence was searched using vmatch v2.3.0 (http://www.vmatch.de/, accessed on 23 March 2023) software, in which parameters were set as minimum length = 30 bp and Hamming distance = 3. Four forms of interspersed repeat sequences were identified: forward, palindromic, reverse and complementary repeat sequences. The SSRs were identified using MISA v1.0 (MIcroSAtellite identification tool, http://pgrc.ipk-gatersleben.de/misa/misa.html, accessed on 23 March 2023) software, in which parameters were set as single-base repeat > 8, di-base repeat > 5, tri-base repeat > 3, tetra-base repeat > 3, penta-base repeat > 3, and hexa-base repeat > 3 [15,39,40]. The SSR primers were designed using the SSR analysis results.
2.6. Cp Genome Comparison of Cabbage Species
Four variants of B. oleracea were selected for comparing the boundaries between the LSC, IR and SSC regions in the kohlrabi cp genome, including the newly assembled cp genome of B. oleracea var. gongylodes (MW900251, 153,364 bp), B. oleracea var. alboglabra cv. SJCT (OR063915, 153,365 bp) [15], B. oleracea var. alboglabra cv. FZHH (OR063916, 153,420 bp) [15] and B. oleracea var. itaica (MN649876.1, 153,364 bp) [23]. Boundary differences in IRB-LSC, IRB-SSC, IRA-SSC, and IRA-LSC were compared among these four variants with the annotation information of cp genomes available in GenBank.
2.7. Phylogenetic Analysis
Phylogenetic relationships were analyzed using MEGA7 by the maximum likelihood (ML) method [41]. The cp genomes of other species for phylogenetic relationships analysis were downloaded from the NCBI database, including B. oleracea (wild cabbage) (MG717288), B. oleracea var. capitata (KR233156.1), B. oleracea var. botrytis (KX681655.1), B. oleracea var. italica (MH388765), B. juncea (KT581449.1), B. rapa (NC_040849.1), B. rapa ssp. rapa (MT409177), B. napus (GQ861354.1), B. nigra (KT878383.1), Raphanus staivus (NC_024469.1), Arabidopsis thaliana (NC_000932.1), Solanum lycopersicum (NC_007898.3) and Oryza sativa (NC_031333.1). Of them, the cp genome sequence of O. sativa (NC 031333.1) was used as the outgroup.
3. Results
3.1. Complete Chloroplast Genome Assembly and Gene Annotation
Based on the assembled sequences, the complete quadripartite circular cp genome of kohlrabi with a length of 153,364 bp without any gaps was generated. Similarly to cp genomes of other crops, there are four sequence regions of the kohlrabi cp genome, including a large single-copy region (LSC) with length of 83,136 bp, a small single-copy region (SSC) with length of 17,834 bp and two inverted repeats (IRa and IRb) with length of 26,197 bp. The base composition analysis showed that the overall GC and AT content was 36.36% and 63.64% of the complete cp genome sequence, respectively. The GC and AT content was 34.15%, 65.85% in the LSC region, 29.10%, 70.90% in the SSC region, and 42.35%, 57.65% in the IR regions, respectively. Based on the gene annotation results, the cp genome of kohlrabi contained 132 genes, 87 of which were annotated as protein-coding genes, 37 as tRNA genes, and 8 as rRNA genes. The complete cp genome map is shown in Figure 2. All sequence information and gene annotation of the complete kohlrabi cp genome has been uploaded to the NCBI database under GenBank accession number MW900251.
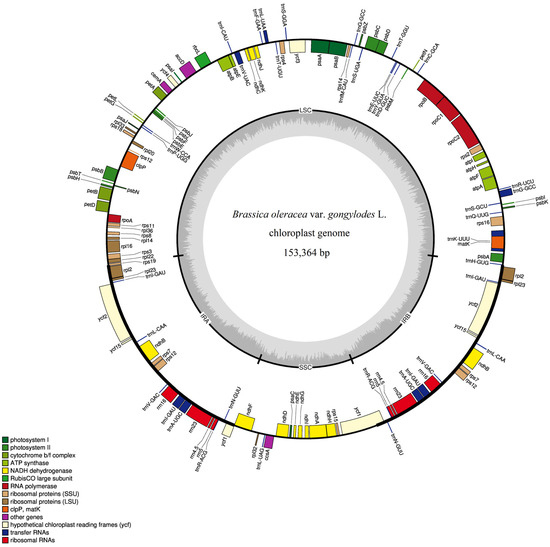
Figure 2.
Quadripartite circular cp genome map of complete kohlrabi chloroplast genome. Note: Genes inside the circle are transcribed clockwise, and genes outside the circle are counterclockwise. The darker gray area and the lighter gray area in the inner circle correspond to GC content and AT content, respectively.
According to the different biosynthetic pathways and various functions of these 132 genes, 45 of them were annotated as involved in photosynthesis pathways, 74 as self-replication genes, 8 considered conserved hypothetical chloroplast open reading frames, and 5 were annotated as other genes (Table 1). Among these 45 genes involved in photosynthesis, 5 genes were involved in subunits of photosystem I—psaA, psaB, psaC, psaI and psaJ; 15 in subunits of photosystem II—psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbZ and psbT; 6 in subunits of the cytochrome b/f complex—petA, petB, petD, petG, petL and petN; 1, rbcL, in a large subunit of rubisco; six in subunits of ATP synthase—atpA, atpB, atpE, atpF, atpH and atpI; and 12 in subunits of NADH dehydrogenase—ndhA, ndhB (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ and ndhK. Five genes contained one intron (petB, petD, atpF, ndhA and ndhB).

Table 1.
Structural characteristics of kohlrabi chloroplast genes.
Among these 74 genes annotated as self-replication genes, 8 were annotated as rRNA genes, comprising rrn16 (×2), rrn23 (×2), rrn4.5 (×2), and rrn5 (×2). Fourteen genes were annotated as involving the small ribosome subunit: rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 (×2), rps14, rps15, rps16, rps18 and rps19. Eleven genes were annotated as involving the large ribosome subunit: rpl2 (×2), rpl14, rpl16, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, and rpl36. Two genes contained one intron (rps16 and rpl2) and one gene (rps12) contained two introns. Among 37 genes annotated as tRNA genes, seven occurred in two copies (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, RNA-UGC, trnR-ACG and trnN-GUU) and six contained one intron (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU and trnA-UGC).
Five other genes were annotated involving maturase (matK), envelope membrane protein (cemA), subunit of acetyl-CoA (accD), c-type cytochrome synthesis gene (ccsA) and protease (clpP), respectively. Of these, clpP was found to contain two introns. Eight genes of unknown function were also identified: ycf1 (×2), ycf2 (×2), ycf3, ycf4, and ycf15 (×2). Of these, ycf3 was found to contain two introns.
Among 19 genes with two copies, except for ycf1 residing within the SSC region and two copies of rps12 residing within the LSC region and IR region, 17 were all located in the IR regions: ndhB, rrn4.5, rrn5, rrn16, rrn23, rps7, rps12, rpl2, rpl23, trnI-CAU, trnV-GAC, trnL-CAA, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU, ycf1, ycf2 and ycf15.
3.2. Relative Synonymous Codon Usage Analysis
Based on the preference of codons used by CDS, we estimated the relative synonymous codon usage frequency (RSCU) and codon usage frequency. The codon–anticodon recognition patterns of the kohlrabi cp genome showed that a total of 30 tRNAs comprised codons corresponding to all 20 essential amino acids for protein biosynthesis. A total of 65 kinds of codons were searched in the cp genome, of which UAA had the highest usage encoding the termination codon. In addition, UUA for leucine, AUG for methionine, GCU for alanine, AGA for arginine, UCU for serine and GGA for glycine also had high usage (Figure 3, Table S1). Moreover, of all these 65 codons, 33 codons had RSCU values of >1, and 29 of them (93.50%) ended with base A or U, whereas the RSCU values for 31 codons were <1, and 30 of them (90.90%) ended with base C or G. Trp was encoded by only one UGG codon, indicating no biased usage (RSCU = 1).
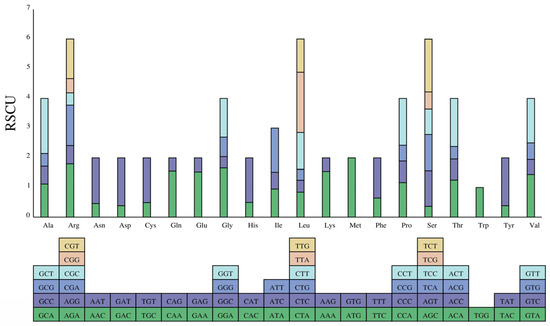
Figure 3.
Analysis of codon preference in the kohlrabi chloroplast genome. Note: The colored blocks represent all codons encoding each amino acid, and the height of the upper column represents the sum of all codon RSCU values.
3.3. Interspersed Repeat Sequence Analysis
Interspersed repeat sequence analysis identified a total of 35 scattered repeats: 11 forward, 21 palindromic, and 3 IRs (Figure 4). The positions of these interspersed repeat sequences were analyzed, and 21, 4, 2 and 7 were distributed in the LSC, SSC, IRa and IRb regions, respectively (Table S2). Repeat sequence lengths ranged from 30 to 47 bp, except the IR region.
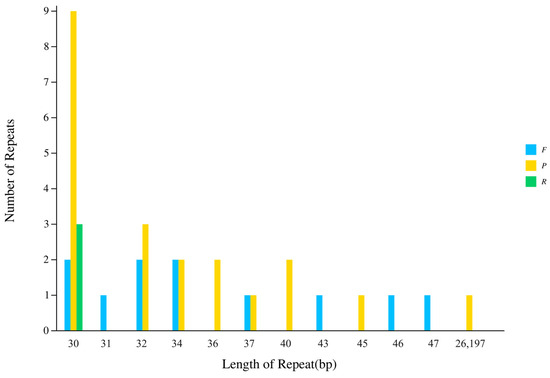
Figure 4.
Summary of interspersed repeat sequence. F represents forward repeat, P represents palindrome repeat, R represents inverted repeat, and C represents complementary repeat.
3.4. Simple Sequence-Repeat Analysis
The SSR analysis revealed that 290 SSRs were identified, including 205 single-base, 18 di-base, 62 tri-base and five tetra-base repeats (Figure 5a). The positions of these SSRs were analyzed, and 188 (64.83%), 60 (20.69%) and 42 (14.48%) were distributed in LSC, SSC and IR regions, respectively (Figure 5b). A total of 98 (33.79%), 42 (14.48%) and 150 (51.72%) SSRs were located in exons, introns, and intergenic regions of the genome (Table S3). Based on the SSR analysis result, 283 pairs of primers were designed (Table S4).
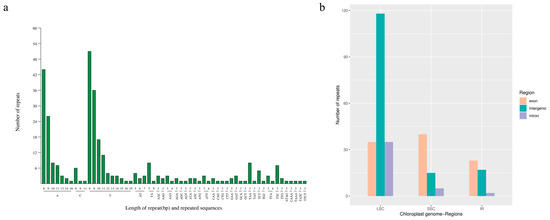
Figure 5.
Summary and distribution of 290 simple sequence repeats. (a) SSR statistical data. (b) SSR distribution.
3.5. Boundary Analysis
Four B. oleracea varieties were selected for boundary analysis between the LSC, IR and SSC regions in the cp genome: the newly assembled cp genome of B. oleracea var. gongylodes (MW900251, 153,364 bp), B. oleracea var. alboglabra cv. SJCT (OR063915, 153,365 bp) [15], B. oleracea var. alboglabra cv. FZHH (OR063916, 153,420 bp) [15] and B. oleracea var. itaica (MN649876.1, 153,364 bp) [23] (Figure 6). The lengths of the IR and SSC regions were the same in these four B. oleracea varieties. Only the length of LSC region was different, and ranged from 83,136 to 83,192 bp. The rps19 coding sequence was located in the boundary of the LSC and IRb region and the relative location was same in these four B. oleracea varieties at 113 bp upstream of the IRb region. The ndhF and ycf1 coding sequences were located in the boundaries of the IRb and SSC region and SSC and IRa region at 2204 bp upstream of the SSC region and 1027 bp upstream of the IRa region, respectively. The tRNA non-coding gene trnH-GUG in these four B. oleracea varieties was within the LSC region, which started 3 bp upstream of LSC in B. oleracea var. gongylodes, B. oleracea var. alboglabra cv. SJCT and B. oleracea var. alboglabra cv. FZHH, and 4 bp upstream of LSC in B. oleracea var. itaica. These results suggested that the boundaries between the LSC, IR and SSC regions were highly conserved except for minor differences in distance of trnH-GUG at the boundary between the IRa and LSC regions in B. oleracea varieties.
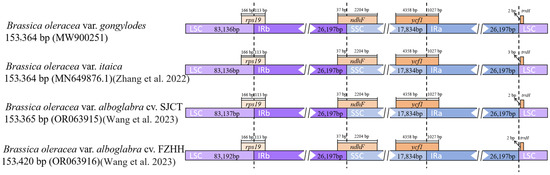
Figure 6.
Comparison of gene location in LSC, IR, and SSC boundaries among four chloroplast genomes of Brassica oleracea varieties [15,23].
3.6. Phylogenetic Relationship Analysis
Phylogenetic relationships among eleven Brassica species, one S. lycopersicum, one O. sativa, and kohlrabi were determined by MEGA7 using the maximum likelihood (ML) method. Of these cp genome, O. sativa (NC_031333.1) was used as an outgroup (Figure 7). Five B. oleracea varieties were clustered into one group and three of them were clustered into one subgroup—B. oleracea var. Botrytis (KX681665.1), B. oleracea var. gongylodes and B. oleracea var. italica (MH388765)—in which the kohlrabi cp genome was closely related to B. oleracea var. botrytis. These results may provide new insight for understanding the polyploidization between Brassicaceae species.
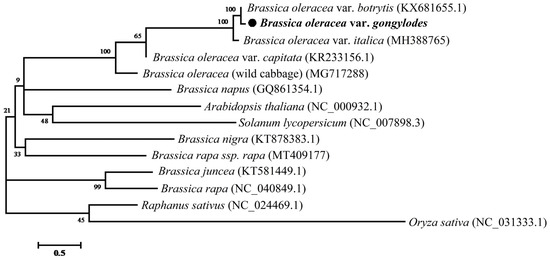
Figure 7.
Phylogenetic relationships of thirteen species and kohlrabi based on the complete chloroplast genome sequence. The chloroplast sequence of O. sativa (NC 031333.1) was used as the outgroup. B. oleracea (wild cabbage) (MG717288), B. oleracea var. capitata (KR233156.1), B. oleracea var. botrytis (KX681655.1), B. oleracea var. italica (MH388765), B. juncea (KT581449.1), B. rapa (NC_040849.1), B. rapa ssp. rapa (MT409177), B. napus (GQ861354.1), R. staivus (NC_024469.1), B. nigra (KT878383.1), A. thaliana (NC_000932.1), S. lycopersicum (NC_007898.3) and O. sativa (NC_031333.1).
4. Discussion
Chloroplasts act as uniparentally inherited semi-autonomous organelles involved in the genetic systems of plants. Zhang et al. [23], Timmis et al. [41] and Liu et al. [42] reported that the gene number, gene composition, and gene arrangement of cp genomes are more highly conserved than those of mitochondrial and nuclear genomes. Based on the cp genome assembly, the length of the kohlrabi cp genome and GC content were similar to those of other Brassicaceae species, such as B. oleracea var. alboglabra [15], B. oleracea var. italica [23], B. rapa ssp. rapa [43], R. sativus L. [44], B. nigra and B. oleracea [45] and B. juncea (Indian mustard) [46]. The cp genome length of these species was approximately 153,300 bp to 153,500 bp and the GC content was approximately 36.30%. In terms of length of cp genome and GC content, they were highly conserved.
A total of 132 genes were identified and annotated in the cp genome of kohlrabi, fewer than that in B. oleracea var. alboglabra and B. oleracea var. itaica. Based on the gene annotation, 19 genes were identified with two copies in the IR regions. In addition, five orf genes of unknown function were also identified in the cp genome of kohlrabi, fewer than that in cpDNA of A. thaliana [47]. This result indicated that gene losses occurred in the cp genome of Brassicaceae family, which is similar to gene losses in the cp genome of other genera, such as Asteraceae, Leguminosae and Gentianaceae [48,49].
Codon usage frequency is a crucial factor influencing the evolution of the cp genome. According to the RSCU estimation, we found that most codons with RSCU values > 1 ended with A or U, while most codons with RSCU values < 1 ended with C or G. This result is consistent with B. oleracea var. itaica [23], Magnoliz zenii [50] and other species [51,52], suggesting that this phenomenon may be similar in plant cp genomes and codon usage frequency of the cp genome is also highly conserved.
In the cp genome of kohlrabi, 290 SSRs were identified, of which 205 (70.69%) belonged to single-base A or T repeats. The proportion of mononucleotide repeats among all SSRs of the kohlrabi cp genome was similar to that in B. oleracea var. itaica [23], B. rapa ssp. rapa [43], Quercus acutissima [53] and Aristolochia medicinal species [54]. Similarly to other reports, most identified SSRs were positioned at the intergenic region of the cp genome. A total of 283 pairs of primers designed relying on SSRs in the cp genome of kohlrabi could be used in DNA fingerprint development and phylogenetic discrepancy analysis.
5. Conclusions
The complete cp genome of kohlrabi with length of 153,364 bp without any gaps was sequenced and analyzed. In sum, 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes were annotated. The overall GC content was 36.36% of the complete cp genome sequence, and 35 scattered repeats and 290 SSRs were found and identified. Phylogenetic relationship analysis revealed that the kohlrabi chloroplast genome was closely related to that of B. oleracea var. botrytis. Our results provide a basis for understanding the chloroplast-dependent metabolic studies and provide new insights into the polyploidization of Brassicaceae species.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes15050550/s1 Table S1 Codon preference analysis data. Table S2. Interspersed repeat sequence data and distribution. Table S3. Simple sequence-repeat data and distribution. Table S4. SSR primer sequence list based on the kohlrabi chloroplast genome sequence.
Author Contributions
Conceptualization, methodology, software, formal analysis, and writing—original draft preparation, M.Z.; software, validation, investigation, data curation, and writing—original draft preparation, Y.W.; Conceptualization, methodology, resources, data curation, supervision, writing—original draft preparation, and writing—review and editing, Y.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a project of the Qinghai Provincial Key Laboratory of Vegetable Genetics and Physiology (4 January 2023) and the Qinghai Province High-End Innovative Talents Plan (K9923197).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MW900251, accessed on 23 March 2023, under accession number MW900251. The associated BioProject, SRA, and BioSample numbers are PRJNA721268, SRR14211765 and SAMN18713453.
Acknowledgments
The authors would like to thank Shaanxi Breeding Biotechnologies Co., Ltd., for DNA sequencing and genome assembly. All authors have consented to the acknowledgement.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lim, T. Brassica oleracea (gongylodes group). In Edible Medicinal and Non Medicinal Plants; Springer: Dordrecht, The Netherlands, 2015; pp. 768–776. [Google Scholar]
- Prajapati, R.; Seong, S.H.; Kim, H.R.; Jung, H.A.; Choi, J.S. Isolation and Identification of Bioactive Compounds from the Tuber of Brassica oleracea var. gongylodes. Nat. Prod. Sci. 2020, 26, 214–220. [Google Scholar] [CrossRef]
- Golob, A.; Novak, T.; Maršić, N.K.; Šircelj, H.; Stibilj, V.; Jerše, A.; Kroflič, A.; Germ, M. Biofortification with selenium and iodine changes morphological properties of Brassica oleracea L. var. gongylodes) and increases their contents in tubers. Plant Physiol. Biochem. 2020, 150, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Jahangir, M.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Health-affecting compounds in Brassicaceae. Compr. Rev. Food Sci. Food Saf. 2009, 8, 31–43. [Google Scholar] [CrossRef]
- Park, W.T.; Kim, J.K.; Park, S.; Lee, S.-W.; Li, X.; Kim, Y.B.; Uddin, M.R.; Park, N.I.; Kim, S.-J.; Park, S.U. Metabolic profiling of glucosinolates, anthocyanins, carotenoids, and other secondary metabolites in kohlrabi (Brassica oleracea var. gongylodes). J. Agric. Food Chem. 2012, 60, 8111–8116. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Z.; Zhu, M.; Zhu, Z.; Wang, Z.; Tian, S.; Chen, G. Anthocyanin accumulation and molecular analysis of correlated genes in purple kohlrabi (Brassica oleracea var. gongylodes L.). J. Agric. Food Chem. 2015, 63, 4160–4169. [Google Scholar] [CrossRef] [PubMed]
- Vue, M.; Rydberg, M.; Talaga, M.L. Purification of a Plant Lectin from Kohlrabi/Brassica oleracea var. gongylodes and Identification of Binding Specificity. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Krzysztof, B.; Burch-Smith, T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015, 6, 781. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M. The chloroplast genome. Plant Mol. Biol. 1992, 19, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Margulis, L. The Origin of eukaryotic cells. Communication 2007, 59, 219–223. [Google Scholar]
- Cavalier-Smith, T. The phagotrophic origin of eukaryotes and phylogenetic classifi cation of Protozoa. Int. J. Syst. Evol. Microbiol. 2002, 52, 297–354. [Google Scholar] [CrossRef]
- Chung, H.Y.; Won, S.Y.; Kim, Y.K.; Kim, J.S. Development of the chloroplast genome-based InDel markers in Niitaka (Pyrus pyrifolia) and its application. Plant Biotechnol. Rep. 2019, 13, 51–61. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.H.; Hong, W.J.; Jung, K.H. A Systematic View Exploring the Role of Chloroplasts in Plant Abiotic Stress Responses. BioMed Res. Int. 2019, 18, 6534745. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, Q.; Zhang, C.; Huang, H.; He, H.; Wang, M.; Li, M.; Huang, Z.; Tang, Y.; Chen, Q.; et al. Sequencing and Analysis of Complete Chloroplast Genomes Provide Insight into the Evolution and Phylogeny of Chinese Kale (Brassica oleracea var. alboglabra). Int. J. Mol. Sci. 2023, 24, 10287. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Biggs, P.J.; Matthews, P.J.; Collins, L.J.; Hendy, M.D.; Lockhart, P.J. Mutational dynamics of aroid chloroplast genomes. Genome Biol. Evol. 2012, 4, 1316–1323. [Google Scholar] [CrossRef]
- Sloan, D.B.; Triant, D.A.; Forrester, N.J.; Bergner, L.M.; Wu, M.; Taylor, D.R. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae). Mol. Phylogenetics Evol. 2014, 72, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Wickett, N.J.; Forrest, L.L.; Budke, J.M.; Shaw, B.; Goffinet, B. Frequent pseudogenization and loss of the plastid-encoded sulfate-transport gene cysA throughout the evolution of liverworts. Am. J. Bot. 2011, 98, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, D.J.; Bendich, A.J. The linear plastid chromosomes of maize: Terminal sequences, structures, and implications for DNA replication. Curr. Genet. 2016, 62, 431–442. [Google Scholar] [CrossRef]
- Duan, H.; Guo, J.; Xuan, L.; Wang, Z.; Li, M.; Yin, Y.; Yang, Y. Comparative chloroplast genomics of the genus Taxodium. BMC Genom. 2020, 21, 114. [Google Scholar] [CrossRef]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The Complete Chloroplast Genome Sequence of Pelargonium × hortorum: Organization and Evolution of the Largest and Most Highly Rearranged Chloroplast Genome of Land Plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tao, M.; Shan, X.; Pan, Y.; Sun, C.; Song, L.; Pei, X.; Jing, Z.; Dai, Z. Characterization of the complete chloroplast genome of Brassica oleracea var. italica and phylogenetic relationships in Brassicaceae. PLoS ONE 2022, 17, e0263310. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, D.A. Complete chloroplast genome of Abutilon fruticosum: Genome structure, comparative and phylogenetic analysis. Plants 2021, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Tian, M.; Zhang, G.; Shao, D.; Ren, Y. Complete chloroplast genome sequence of turnip (Brassica rapa. ssp. rapa): Genome structure and phylogenetic analysis. Mitochondrial DNA Part B 2020, 5, 3573–3575. [Google Scholar] [CrossRef]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecular 2018, 23, 246. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Nie, L.; Sun, W.; Xu, Z.; Wang, Y.; Yu, J.; Song, J.; Yao, H. Comparative and Phylogenetic Analyses of Ginger (Zingiber officinale) in the Family Zingiberaceae Based on the Complete Chloroplast Genome. Plants 2019, 8, 283. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, F.; Abdullah, S.I.; Ahmed, I.; Waheed, M.T.; Mirza, B. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics 2020, 112, 1522–1530. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Nguyen, T.N.L.; Doan, T.N.; Phạm, M.H.; Le, T.L.; Sy, D.T.; Chu, H.H. Complete chloroplast genome of novel Adrinandra megaphylla Hu species: Molecular structure, comparative and phylogenetic analysis. Sci. Rep. 2021, 11, 11731. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Wen, J.; Guo, H.; Liu, Y. Comparative and phylogenetic analyses based on the complete chloroplast genome of Cornus subg. Syncarpea (Cornaceae) species. Front. Plant Sci. 2024, 15, 1306196. [Google Scholar] [CrossRef]
- Abdullah; Henriquez, C.L.; Mehmood, F.; Shahzadi, I.; Ali, Z.; Waheed, M.T.; Croat, T.B.; Poczai, P.; Ahmed, I. Comparison of chloroplast genomes among species of unisexual and bisexual clades of the monocot family araceae. Plants 2020, 9, 737. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Shao, D.; Ma, Y.; Li, X.; Ga, S.; Ren, Y. The sequence structure and phylogenetic analysis by complete mitochondrial genome of kohlrabi (Brassica oleracea var. gongylodes L.). Mitochondrial DNA Part B 2021, 6, 2714–2716. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organelle genomes with DOGMA. Bioinformation 2004, 20, 325–3255. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. Organellar genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W5891. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Li, M.; Zhang, R.; Li, J.; Zheng, K.; Xiao, J.; Zheng, Y. Analyses of chloroplast genome of Eutrema japonicum provide new insights into the evolution of Eutrema species. Agronomy 2021, 11, 2546. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Liu, H.Y.; Yu, Y.; Deng, Y.Q.; Li, J.; Huang, Z.X.; Zhou, S.D. The chloroplast genome of Lilium henrici: Genome structure and comparative analysis. Molecules 2018, 23, 1276. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y. The complete mitochondrial genome of turnip (Brassica rapa ssp. rapa). Mitochondrial DNA Part B 2021, 6, 1566–1567. [Google Scholar] [CrossRef]
- Jeong, Y.M.; Chung, W.H.; Mun, J.H.; Kim, N.; Yu, H.J. De novo assembly and characterization of the complete chloroplast genome of radish (Raphanus sativus L.). Gene 2014, 551, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Seol, Y.J.; Kim, K.; Kang, S.H.; Perumal, S.; Lee, J.; Kim, C.K. The complete chloroplast genome of two Brassica species, Brassica nigra and B. oleracea. Mitochondrial DNA Part B 2015, 28, 167–168. [Google Scholar] [CrossRef] [PubMed]
- Prabhudas, S.K.; Raju, B.; Thodi, S.K.; Parani, M.; Natarajan, P. The complete chloroplast genome sequence of Indian mustard (Brassica juncea L.). Mitochondrial DNA 2016, 27, 4622–4623. [Google Scholar] [CrossRef] [PubMed]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Walker, J.F.; Zanis, M.J.; Emery, N.C. Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae). Am. J. Bot. 2014, 101, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.S.; Fu, P.C.; Zhou, X.J.; Cheng, Y.W.; Zhang, F.Q.; Chen, S.L.; Gao, Q.B. The complete plastome sequences of seven species in Gentianasect. Kudoa (Gentianaceae): Insights into plastid gene loss and molecular evolution. Front. Plant Sci. 2018, 9, 493. [Google Scholar] [CrossRef]
- Li, Y.F.; Sylvester, S.P.; Li, M.; Zhang, C.; Li, X.; Duan, Y.; Wang, X. The complete plastid genome of Magnolia zenii and genetic comparison to Magnoliaceae species. Molecular 2019, 24, 261. [Google Scholar] [CrossRef]
- Redwan, R.M.; Saidin, A.; Kumar, S.V. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 2015, 15, 196. [Google Scholar] [CrossRef]
- Li, B.; Lin, F.; Huang, P.; Guo, W.; Zheng, Y. Complete chloroplast genome sequence of decaisnea insignis: Genome organization, genomic resources and comparative analysis. Sci. Rep. 2017, 7, 10073. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Zang, M.; Li, M.; Fang, Y. Complete chloroplast genome sequence and phylogenetic analysis of Quercus acutissima. Int. J. Mol. Sci. 2018, 19, 2443. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zuo, Y.; Zhu, X.; Liao, S.; Ma, J. Complete chloroplast genomes and comparative analysis of sequences evolution among seven Aristolochia (Aristolochiaceae) medicinal species. Int. J. Mol. Sci. 2019, 20, 1045. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).