LncRNA NDUFA6-DT: A Comprehensive Analysis of a Potential LncRNA Biomarker and Its Regulatory Mechanisms in Gliomas
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
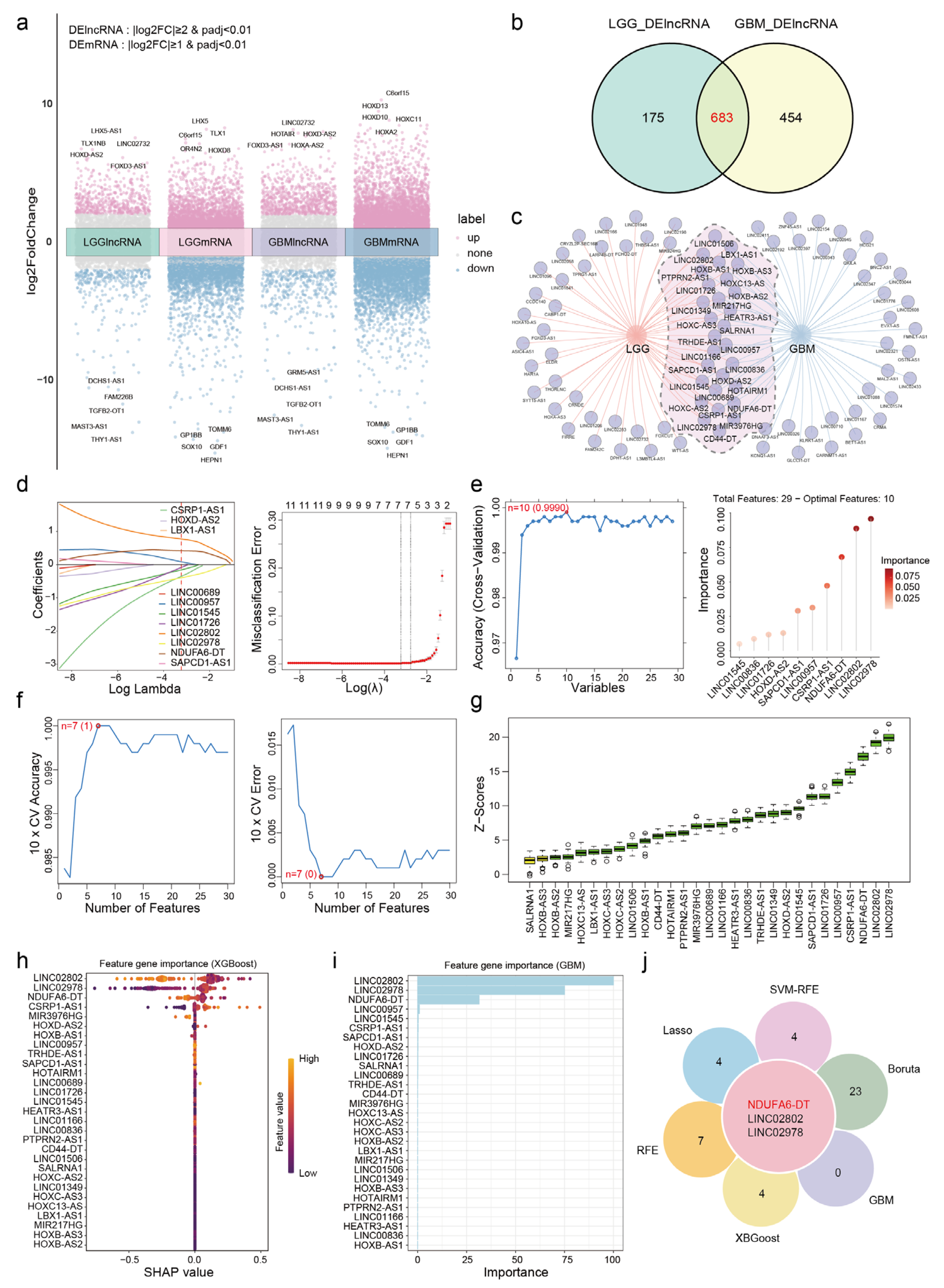
2.2. Identification of Differentially Expressed Genes
2.3. Selection and Validation of Key lncRNAs
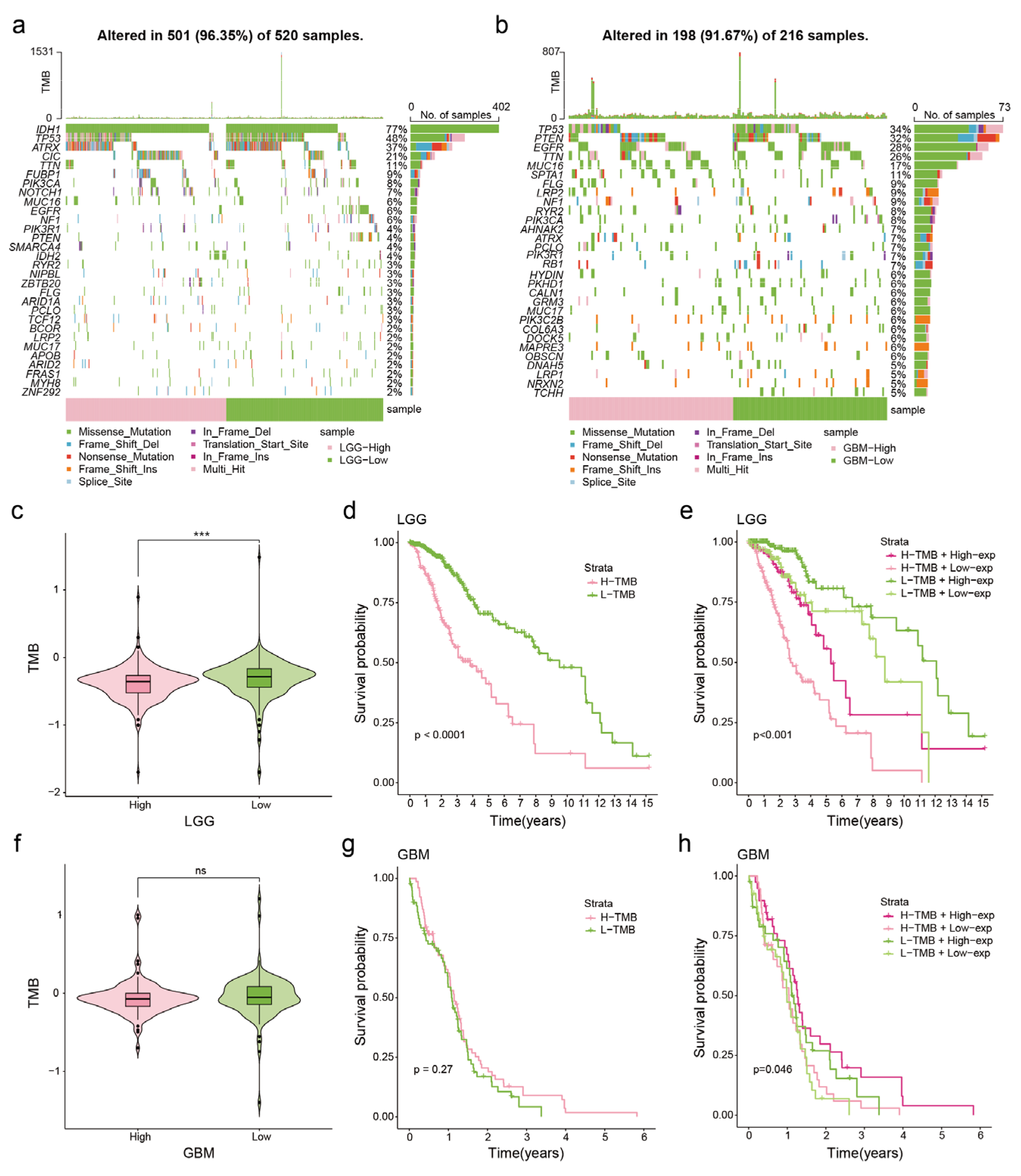
2.4. Somatic Mutation Analysis
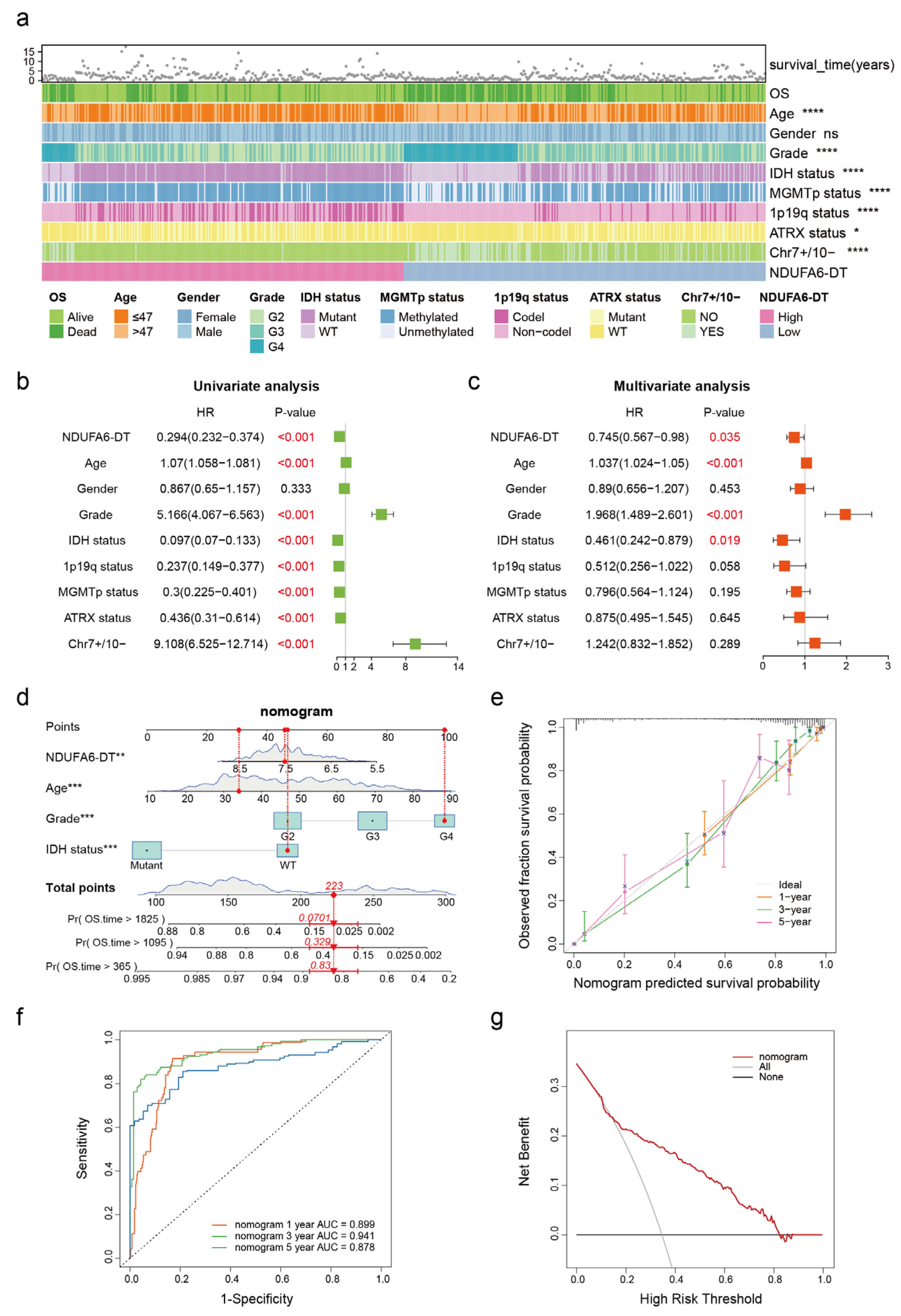
2.5. Construction and Validation of Nomogram
2.6. Pathway Enrichment and Functional Analysis
2.7. Immune Infiltration Analysis
2.8. Prediction of the ceRNA Network for NDUFA6-DT
2.9. Statistical Analysis
3. Results
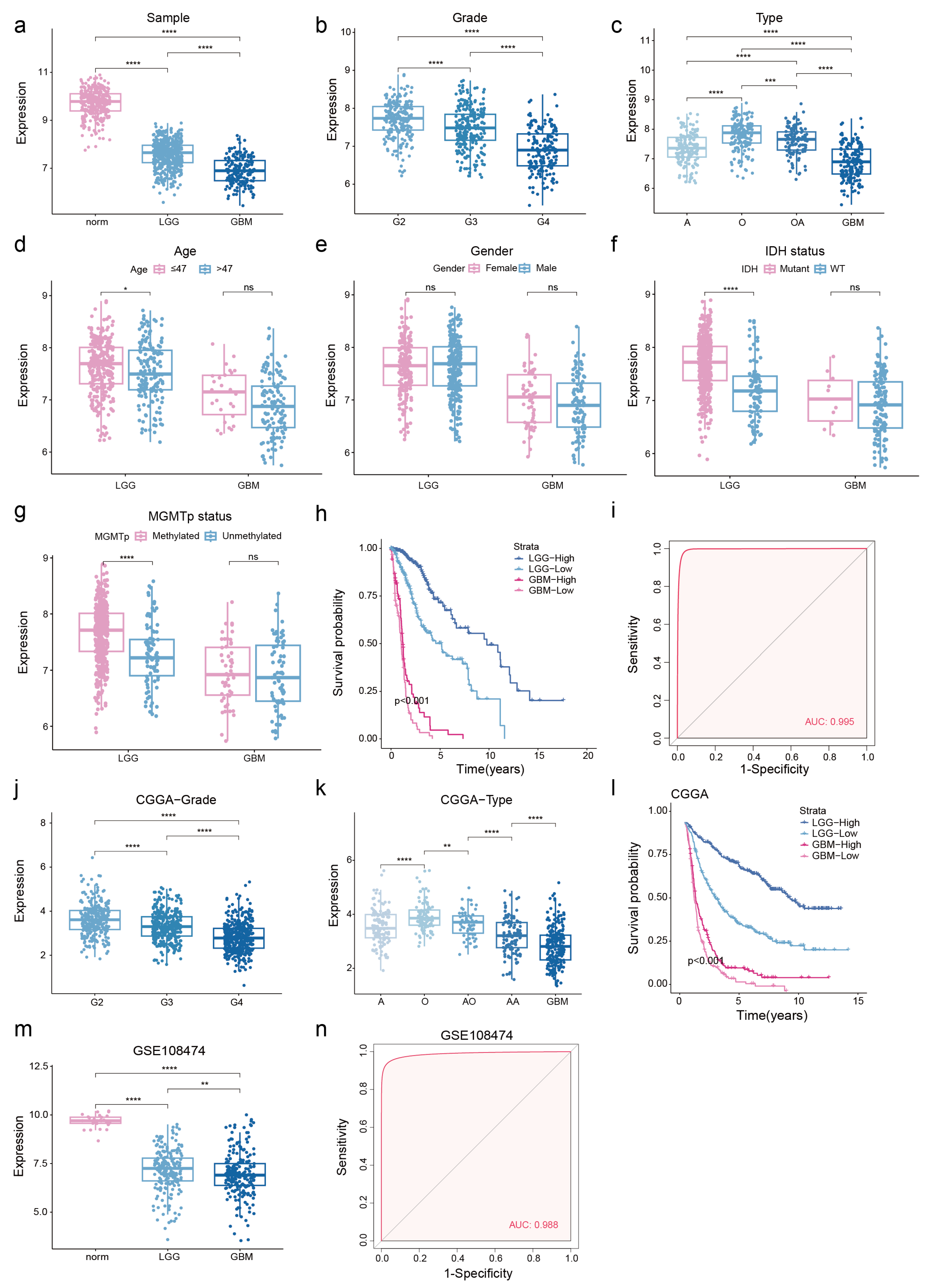
3.1. NDUFA6-DT: A Potential Key lncRNA Signature in Glioma Pathology
3.2. Low NDUFA6-DT Expression in Gliomas Correlates with Adverse Prognosis
3.3. Correlation between NDUFA6-DT Expression and Genetic Mutations in Gliomas
3.4. NDUFA6-DT: An Independent Prognostic Factor and Protective Element in Gliomas
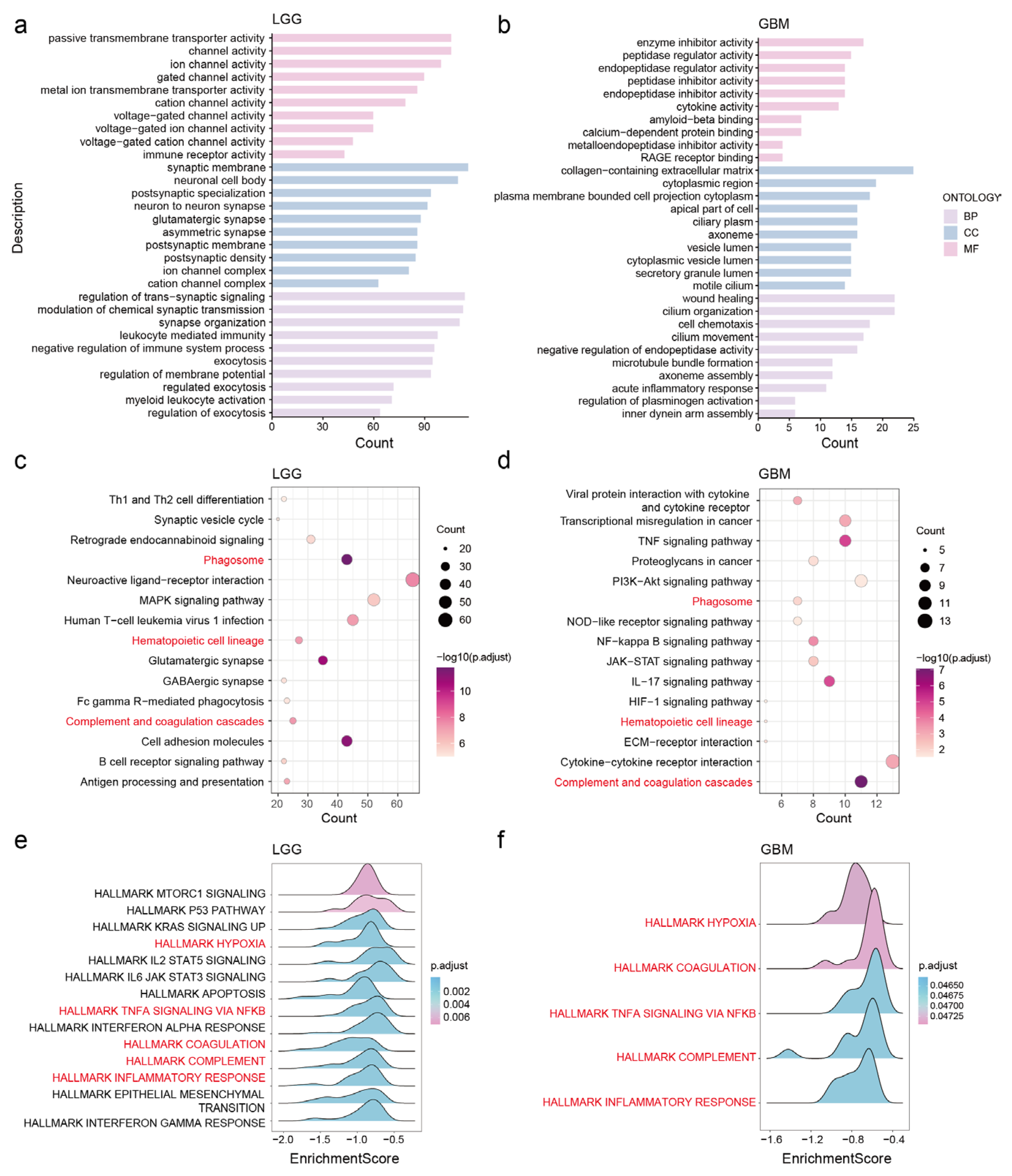
3.5. Potential Functions and Pathways of NDUFA6-DT in Gliomas
3.6. The Expression of NDUFA6-DT Is Associated with Distinct Patterns of Immune Infiltration in Gliomas
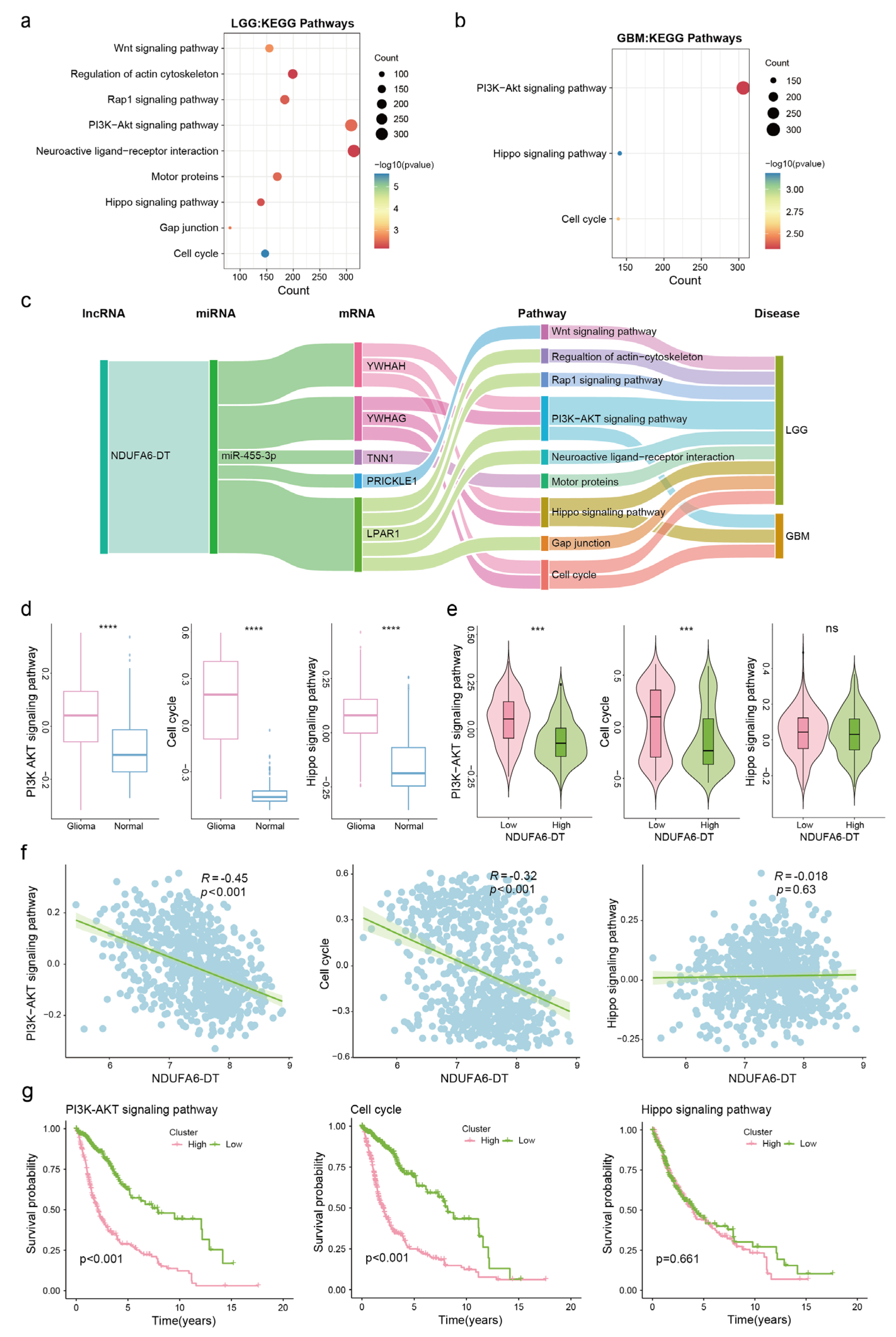
3.7. Potential ceRNA Network of NDUFA6-DT in Gliomas
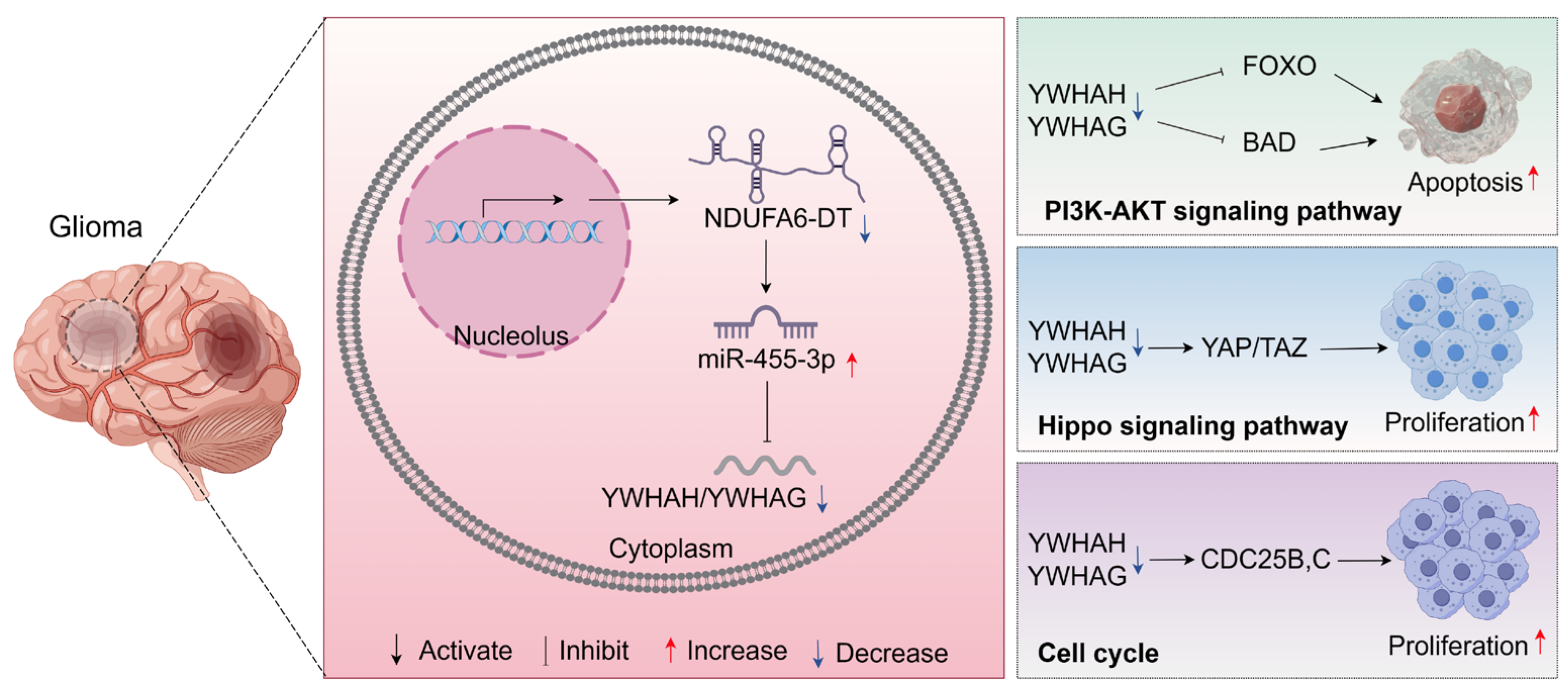
3.8. Potential Regulatory Mechanisms of the NDUFA6-DT-Associated ceRNA Network in Gliomas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Kiran, M.; Chatrath, A.; Tang, X.; Keenan, D.M.; Dutta, A. A Prognostic Signature for Lower Grade Gliomas Based on Expression of Long Non-Coding RNAs. Mol. Neurobiol. 2019, 56, 4786–4798. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, Z.; Wang, X.; Huang, Z.; He, Z.; Chen, Y. Long non-coding RNA: A new player in cancer. J. Hematol. Oncol. 2013, 6, 37. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Kiang, K.M.; Zhang, X.Q.; Leung, G.K. Long Non-Coding RNAs: The Key Players in Glioma Pathogenesis. Cancers 2015, 7, 1406–1424. [Google Scholar] [CrossRef]
- Xi, J.; Sun, Q.; Ma, L.; Kang, J. Long non-coding RNAs in glioma progression. Cancer Lett. 2018, 419, 203–209. [Google Scholar] [CrossRef]
- Xu, C.; He, T.; Li, Z.; Liu, H.; Ding, B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed. Pharmacother. 2017, 95, 1504–1513. [Google Scholar] [CrossRef]
- Zhang, X.; Niu, W.; Mu, M.; Hu, S.; Niu, C. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J. Exp. Clin. Cancer Res. 2020, 39, 196. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Shen, F.; Du, J.; Fang, X.; Li, X.; Su, J.; Wang, X.; Huang, X.; Liu, Z. Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed. Pharmacother. 2018, 97, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gusev, Y.; Bhuvaneshwar, K.; Song, L.; Zenklusen, J.C.; Fine, H.; Madhavan, S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data 2018, 5, 180158. [Google Scholar] [CrossRef]
- Gulluoglu, S.; Tuysuz, E.C.; Sahin, M.; Kuskucu, A.; Kaan Yaltirik, C.; Ture, U.; Kucukkaraduman, B.; Akbar, M.W.; Gure, A.O.; Bayrak, O.F.; et al. Simultaneous miRNA and mRNA transcriptome profiling of glioblastoma samples reveals a novel set of OncomiR candidates and their target genes. Brain Res. 2018, 1700, 199–210. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, J.; Hoadley, K.; Kushwaha, D.; Ramakrishnan, V.; Li, S.; Kang, C.; You, Y.; Jiang, C.; Song, S.W.; et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012, 14, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, M.; Guarnaccia, L.; La Cognata, V.; Navone, S.E.; Campanella, R.; Ampollini, A.; Locatelli, M.; Miozzo, M.; Marfia, G.; Cavallaro, S. A Targeted Next-Generation Sequencing Panel to Genotype Gliomas. Life 2022, 12, 956. [Google Scholar] [CrossRef] [PubMed]
- Ru, Q.; Tian, X.; Pi, M.S.; Chen, L.; Yue, K.; Xiong, Q.; Ma, B.M.; Li, C.Y. Voltage-gated K+ channel blocker quinidine inhibits proliferation and induces apoptosis by regulating expression of microRNAs in human glioma U87-MG cells. Int. J. Oncol. 2015, 46, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Krell, A.; Wolter, M.; Stojcheva, N.; Hertler, C.; Liesenberg, F.; Zapatka, M.; Weller, M.; Malzkorn, B.; Reifenberger, G. MiR-16-5p is frequently down-regulated in astrocytic gliomas and modulates glioma cell proliferation, apoptosis and response to cytotoxic therapy. Neuropathol. Appl. Neurobiol. 2019, 45, 441–458. [Google Scholar] [CrossRef]
- Chen, B.; Lin, P.; Li, N. Targeting the LINC00324/miR-16-5p/SEPT2 Signaling Cascade is Effective to Reverse Malignant Phenotypes in Glioblastoma. Anticancer. Agents Med. Chem. 2023, 23, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Ujifuku, K.; Mitsutake, N.; Takakura, S.; Matsuse, M.; Saenko, V.; Suzuki, K.; Hayashi, K.; Matsuo, T.; Kamada, K.; Nagata, I.; et al. miR-195, miR-455-3p and miR-10a* are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010, 296, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Tezcan, G.; Tunca, B.; Bekar, A.; Preusser, M.; Berghoff, A.S.; Egeli, U.; Cecener, G.; Ricken, G.; Budak, F.; Taskapilioglu, M.O.; et al. microRNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell Mol. Neurobiol. 2014, 34, 679–692. [Google Scholar] [CrossRef]
- Wang, W.; Mu, S.; Zhao, Q.; Xue, L.; Wang, S. Identification of differentially expressed microRNAs and the potential of microRNA-455-3p as a novel prognostic biomarker in glioma. Oncol. Lett. 2019, 18, 6150–6156. [Google Scholar] [CrossRef]
- Lin, L.; Cai, J.; Jiang, C. Recent Advances in Targeted Therapy for Glioma. Curr. Med. Chem. 2017, 24, 1365–1381. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Lou, K.; Liu, H. A novel prognostic signature based on N7-methylguanosine-related long non-coding RNAs in breast cancer. Front. Genet. 2022, 13, 1030275. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Tao, K.; Cai, X. Necroptosis-Associated lncRNA Prognostic Model and Clustering Analysis: Prognosis Prediction and Tumor-Infiltrating Lymphocytes in Breast Cancer. J. Oncol. 2022, 2022, 7099930. [Google Scholar] [CrossRef]
- Gao, S.; Wang, Y.; Xu, Y.; Liu, S. An Angiogenesis-Related lncRNA Signature Is Associated with Prognosis and Tumor Immune Microenvironment in Breast Cancer. J. Pers. Med. 2023, 13, 513. [Google Scholar] [CrossRef]
- Rao, Y.; Liu, H.; Yan, X.; Wang, J. In Silico Analysis Identifies Differently Expressed lncRNAs as Novel Biomarkers for the Prognosis of Thyroid Cancer. Comput. Math. Methods Med. 2020, 2020, 3651051. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, D. Identification of Novel lncRNA Markers in Glioblastoma Multiforme and Their Clinical Significance: A Study Based on Multiple Sequencing Data. Onco Targets Ther. 2020, 13, 1087–1098. [Google Scholar] [CrossRef]
- Niu, X.; Sun, J.; Meng, L.; Fang, T.; Zhang, T.; Jiang, J.; Li, H. A Five-lncRNAs Signature-Derived Risk Score Based on TCGA and CGGA for Glioblastoma: Potential Prospects for Treatment Evaluation and Prognostic Prediction. Front. Oncol. 2020, 10, 590352. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Sasaki, H.; Miwa, T.; Ohba, S.; Ikeda, E.; Abe, M.; Ikeda, S.; Kobayashi, M.; Kawase, T.; Hasegawa, M.; et al. Whole genome analysis from microdissected tissue revealed adult supratentorial grade II-III gliomas are divided into clinically relevant subgroups by genetic profile. Neurosurgery 2011, 69, 376–390. [Google Scholar] [CrossRef]
- Vigneswaran, K.; Neill, S.; Hadjipanayis, C.G. Beyond the World Health Organization grading of infiltrating gliomas: Advances in the molecular genetics of glioma classification. Ann. Transl. Med. 2015, 3, 95. [Google Scholar] [CrossRef]
- Siegal, T. Clinical Relevance of Prognostic and Predictive Molecular Markers in Gliomas. Adv. Tech. Stand. Neurosurg. 2016, 43, 91–108. [Google Scholar] [CrossRef]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Joudi Mashhad, M.; Hassanian, S.M.; Anvari, K.; Avan, A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J. Cell Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.K.; Zhu, X.; Gagea, M.; White, C.L., 3rd; Cote, G.; Georgescu, M.M. PHLPP2 suppresses the NF-kappaB pathway by inactivating IKKbeta kinase. Oncotarget 2014, 5, 815–823. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Y.; Wang, K.; Zhao, Z.; Sun, S.; Zhang, K.; Huang, R.; Zeng, F.; Hu, H. ADAR3 expression is an independent prognostic factor in lower-grade diffuse gliomas and positively correlated with the editing level of GRIA2(Q607R). Cancer Cell Int. 2018, 18, 196. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, X.; Gao, L.; Geng, R.; Tao, X.; Xu, H.; Chen, Z. Expression and Prognostic Role of Glia Maturation Factor-γ in Gliomas. Front. Mol. Neurosci. 2022, 15, 906762. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, X.; Zhu, X.L.; Bai, H.; Wang, Z.Z.; Zhang, J.J.; Hao, C.Y.; Duan, H.B. S100A gene family: Immune-related prognostic biomarkers and therapeutic targets for low-grade glioma. Aging 2021, 13, 15459–15478. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Leng, W.; Chen, J.; Li, S.; Lei, B.; Zhang, H.; Zhao, H. Identification of a copper metabolism-related gene signature for predicting prognosis and immune response in glioma. Cancer Med. 2023, 12, 10123–10137. [Google Scholar] [CrossRef]
- Filppu, P.; Tanjore Ramanathan, J.; Granberg, K.J.; Gucciardo, E.; Haapasalo, H.; Lehti, K.; Nykter, M.; Le Joncour, V.; Laakkonen, P. CD109-GP130 interaction drives glioblastoma stem cell plasticity and chemoresistance through STAT3 activity. JCI Insight 2021, 6, e141486. [Google Scholar] [CrossRef]
- Tong, F.; Zhao, J.X.; Fang, Z.Y.; Cui, X.T.; Su, D.Y.; Liu, X.; Zhou, J.H.; Wang, G.X.; Qiu, Z.J.; Liu, S.Z.; et al. MUC1 promotes glioblastoma progression and TMZ resistance by stabilizing EGFRvIII. Pharmacol. Res. 2023, 187, 106606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, Q.; Zhang, G.; Cao, X.; Chen, W.; Li, Y.; Guan, M.; Yu, J.; Wang, X.; Yan, Y. High myosin binding protein H expression predicts poor prognosis in glioma patients. Sci. Rep. 2022, 12, 1525. [Google Scholar] [CrossRef]
- Barthel, L.; Hadamitzky, M.; Dammann, P.; Schedlowski, M.; Sure, U.; Thakur, B.K.; Hetze, S. Glioma: Molecular signature and crossroads with tumor microenvironment. Cancer Metastasis Rev. 2022, 41, 53–75. [Google Scholar] [CrossRef]
- Linares, C.A.; Varghese, A.; Ghose, A.; Shinde, S.D.; Adeleke, S.; Sanchez, E.; Sheriff, M.; Chargari, C.; Rassy, E.; Boussios, S. Hallmarks of the Tumour Microenvironment of Gliomas and Its Interaction with Emerging Immunotherapy Modalities. Int. J. Mol. Sci. 2023, 24, 13215. [Google Scholar] [CrossRef]
- Wang, T.; Xu, S.; Zhang, L.; Yang, T.; Fan, X.; Zhu, C.; Wang, Y.; Tong, F.; Mei, Q.; Pan, A. Identification of immune-related lncRNA in sepsis by construction of ceRNA network and integrating bioinformatic analysis. BMC Genom. 2023, 24, 484. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Morrison, D.K. The 14-3-3 proteins: Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009, 19, 16–23. [Google Scholar] [CrossRef]
- Obsilova, V.; Obsil, T. Structural insights into the functional roles of 14-3-3 proteins. Front. Mol. Biosci. 2022, 9, 1016071. [Google Scholar] [CrossRef]
- Wu, H.; Wei, M.; Li, Y.; Ma, Q.; Zhang, H. Research Progress on the Regulation Mechanism of Key Signal Pathways Affecting the Prognosis of Glioma. Front. Mol. Neurosci. 2022, 15, 910543. [Google Scholar] [CrossRef]
- Luo, J.; Junaid, M.; Hamid, N.; Duan, J.J.; Yang, X.; Pei, D.S. Current understanding of gliomagenesis: From model to mechanism. Int. J. Med. Sci. 2022, 19, 2071–2079. [Google Scholar] [CrossRef]
- Wang, H.; Zhi, H.; Ma, D.; Li, T. MiR-217 promoted the proliferation and invasion of glioblastoma by repressing YWHAG. Cytokine 2017, 92, 93–102. [Google Scholar] [CrossRef]
- Park, G.Y.; Han, J.Y.; Han, Y.K.; Kim, S.D.; Kim, J.S.; Jo, W.S.; Chun, S.H.; Jeong, D.H.; Lee, C.W.; Yang, K.; et al. 14-3-3 eta depletion sensitizes glioblastoma cells to irradiation due to enhanced mitotic cell death. Cancer Gene Ther. 2014, 21, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.Q.; Zhang, X.; Zhen, Y.; He, X.Y.; Zhao, S.; Li, X.F.; Yang, B.; Gao, F.; Guo, F.Y.; Fu, L.; et al. A novel small-molecule activator of Sirtuin-1 induces autophagic cell death/mitophagy as a potential therapeutic strategy in glioblastoma. Cell Death Dis. 2018, 9, 767. [Google Scholar] [CrossRef]



| Accession/Cohort | Database | RNA Library | Sample Size |
|---|---|---|---|
| TCGA-LGG | TCGA | RNA sequencing | LGG: 528 |
| TCGA-GBM | TCGA | RNA sequencing | GBM: 168 |
| GTEx | GTEx | RNA sequencing | Normal brain: 289 |
| mRNAseq_693 | CGGA | RNA sequencing | LGG: 443, GBM: 249 |
| mRNAseq_325 | CGGA | RNA sequencing | LGG: 182, GBM: 139 |
| GSE108474 | GEO | RNA sequencing | Normal: 28, LGG: 215, GBM: 228 |
| GSE90603 | GEO | miRNA sequencing | Normal: 9, GBM: 16 |
| GSE25631 | GEO | miRNA sequencing | Normal: 5, GBM: 82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, R.; Kong, Y.; Luo, Z.; Li, Q. LncRNA NDUFA6-DT: A Comprehensive Analysis of a Potential LncRNA Biomarker and Its Regulatory Mechanisms in Gliomas. Genes 2024, 15, 483. https://doi.org/10.3390/genes15040483
Huang R, Kong Y, Luo Z, Li Q. LncRNA NDUFA6-DT: A Comprehensive Analysis of a Potential LncRNA Biomarker and Its Regulatory Mechanisms in Gliomas. Genes. 2024; 15(4):483. https://doi.org/10.3390/genes15040483
Chicago/Turabian StyleHuang, Ruiting, Ying Kong, Zhiqing Luo, and Quhuan Li. 2024. "LncRNA NDUFA6-DT: A Comprehensive Analysis of a Potential LncRNA Biomarker and Its Regulatory Mechanisms in Gliomas" Genes 15, no. 4: 483. https://doi.org/10.3390/genes15040483
APA StyleHuang, R., Kong, Y., Luo, Z., & Li, Q. (2024). LncRNA NDUFA6-DT: A Comprehensive Analysis of a Potential LncRNA Biomarker and Its Regulatory Mechanisms in Gliomas. Genes, 15(4), 483. https://doi.org/10.3390/genes15040483






