Advances in Understanding Fusarium graminearum: Genes Involved in the Regulation of Sexual Development, Pathogenesis, and Deoxynivalenol Biosynthesis
Highlights
- Virulence Factors: Summarized critical secreted proteins and DON synthesis in Fusarium graminearum.
- Sexual Reproduction: Discussed the sexual reproduction of Fusarium graminearum and the important genes during this process.
- Gene Engineering Applications: Discussed the potential gene engineering technologies to prevent and control wheat head blight.
Abstract
1. Introduction
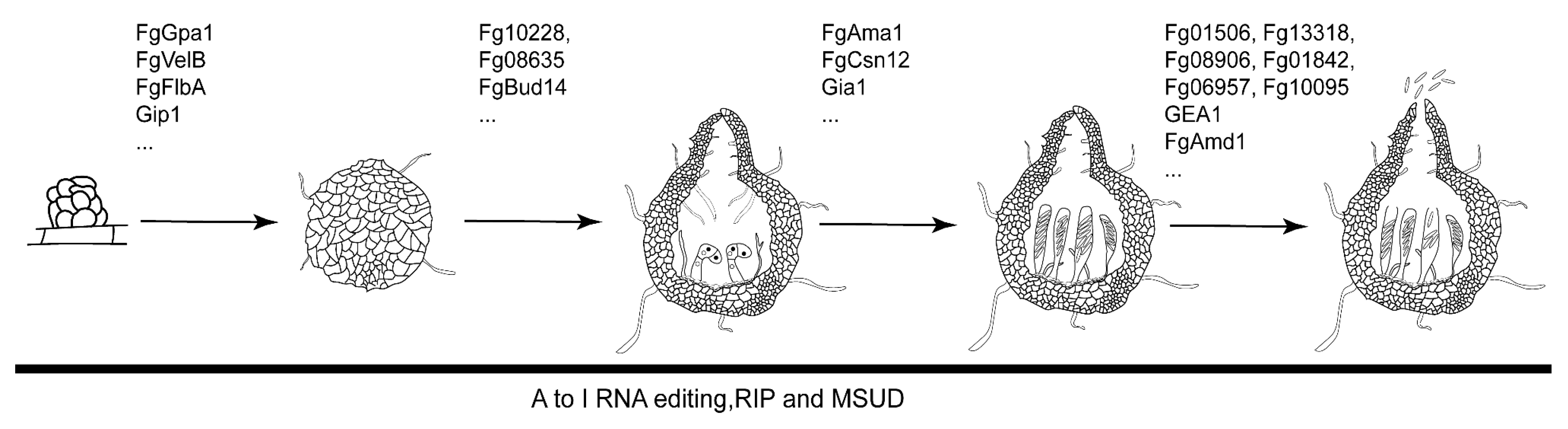
2. Sexual Reproduction of F. graminearum Provides the Primary Inoculum for FHB
2.1. The Sexual Development Processes of F. graminearum
2.2. Genes Involves in the Formation of Perithecia
2.3. Genes Involves in Ascosporogenesis
2.4. Epigenetic Regulation during Sexual Reproduction in F. graminearum
3. Virulence Factors Secreted by F. graminearum during Wheat Infection
3.1. F. graminearum Secretes a Variety of Enzymes and Effectors to Facilitate Infection
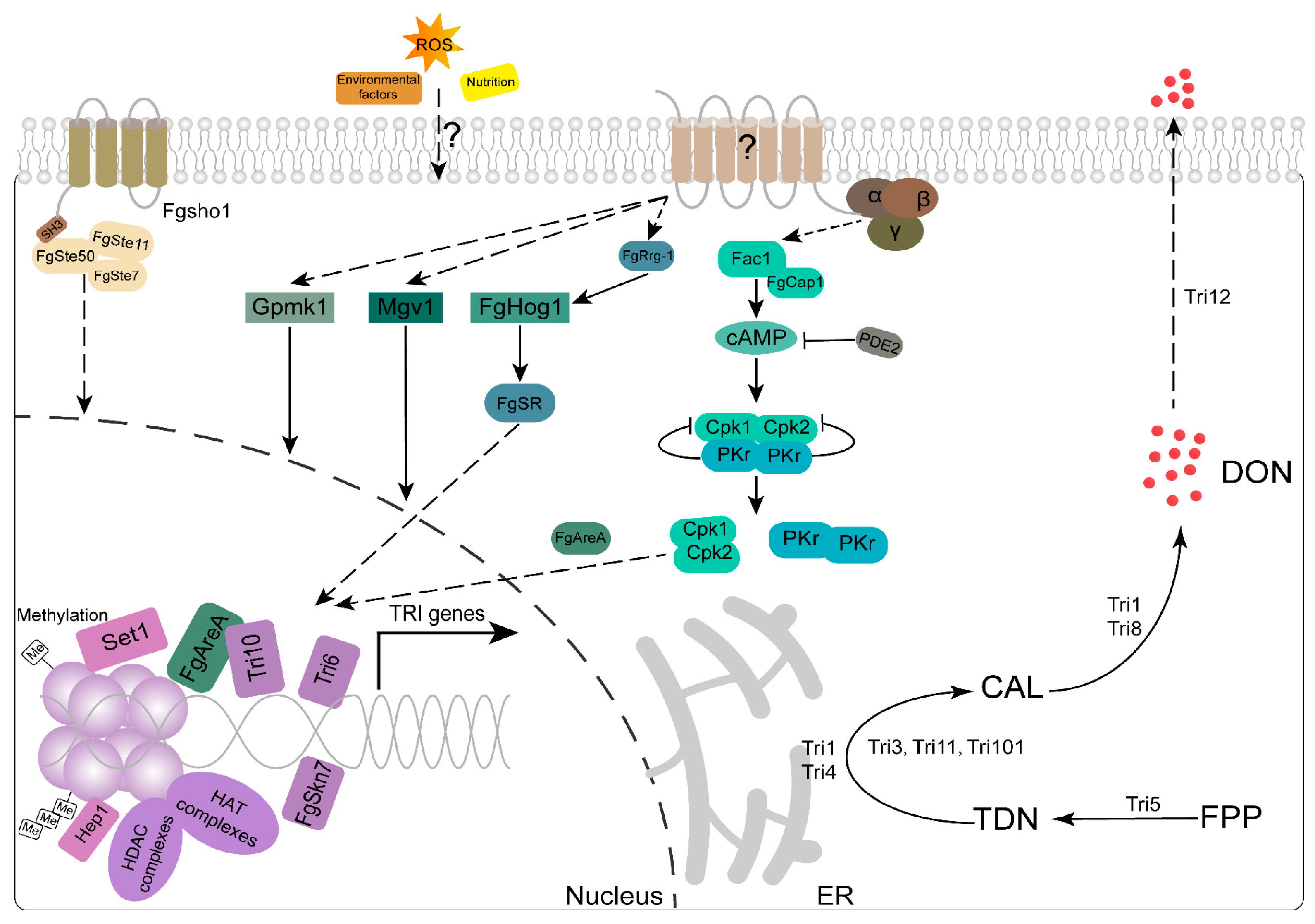
3.2. DON Is a Crucial Virulence Factor Necessary for the Proliferation of Infections on Wheat Heads
3.3. Genes Involves in DON Production
3.4. DON Production and Plant Infection Are Affected by Environment Factors
4. Perspectives
4.1. Disease Control Based on Virulence Gene
4.2. Molecular Design Breeding Based on F. graminearum Effectors
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef]
- Khan, M.K.; Pandey, A.; Athar, T.; Choudhary, S.; Deval, R.; Gezgin, S.; Hamurcu, M.; Topal, A.; Atmaca, E.; Santos, P.A.; et al. Fusarium head blight in wheat: Contemporary status and molecular approaches. 3 Biotech 2020, 10, 172. [Google Scholar] [CrossRef]
- Moonjely, S.; Ebert, M.; Paton-Glassbrook, D.; Noel, Z.A.; Roze, L.; Shay, R.; Watkins, T.; Trail, F. Update on the state of research to manage Fusarium head blight. Fungal Genet. Biol. 2023, 169, 103829. [Google Scholar] [CrossRef]
- Powell, A.J.; Vujanovic, V. Evolution of Fusarium Head Blight Management in Wheat: Scientific Perspectives on Biological Control Agents and Crop Genotypes Protocooperation. Appl. Sci. 2021, 11, 8960. [Google Scholar] [CrossRef]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S.J.M.P.P. A review of wheat diseases—A field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef]
- Ma, Z.; Xie, Q.; Li, G.; Jia, H.; Zhou, J.; Kong, Z.; Li, N.; Yuan, Y. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 2020, 133, 1541–1568. [Google Scholar] [CrossRef]
- Xu, F.; Yang, G.; Wang, J.; Song, Y.; Liu, L.; Zhang, J. Composition and variation in aggressiveness of Fusarium populations causing wheat head blight in Henan province. Phytopathol. Res. 2016, 46, 294–303. [Google Scholar]
- Shah, D.A.; De Wolf, E.D.; Paul, P.A.; Madden, L.V. Predicting Fusarium head blight epidemics with boosted regression trees. Phytopathology 2014, 104, 702–714. [Google Scholar] [CrossRef]
- Trail, F. For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiol. 2009, 149, 103–110. [Google Scholar] [CrossRef]
- Maldonado-Ramirez, S.L.; Schmale III, D.G.; Shields, E.J.; Bergstrom, G.C.J.A.; Meteorology, F. The relative abundance of viable spores of Gibberella zeae in the planetary boundary layer suggests the role of long-distance transport in regional epidemics of Fusarium head blight. Agric. For. Meteorol. 2005, 132, 20–27. [Google Scholar] [CrossRef]
- Quarantin, A.; Castiglioni, C.; Schäfer, W.; Favaron, F.; Sella, L.J.P.P. The Fusarium graminearum cerato-platanins loosen cellulose substrates enhancing fungal cellulase activity as expansin-like proteins. Plant Physiol. Biochem. 2019, 139, 229–238. [Google Scholar] [CrossRef]
- Boenisch, M.J.; Schäfer, W. Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol. 2011, 11, 110. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Q.; Wang, G.; Zhang, X.; Liu, H.; Jiang, C. Combatting Fusarium head blight: Advances in molecular interactions between Fusarium graminearum and wheat. Phytopathol. Res. 2022, 4, 37. [Google Scholar] [CrossRef]
- Brown, N.A.; Urban, M.; van de Meene, A.M.; Hammond-Kosack, K.E. The infection biology of Fusarium graminearum: Defining the pathways of spikelet to spikelet colonisation in wheat ears. Fungal Biol. 2010, 114, 555–571. [Google Scholar] [CrossRef]
- Dweba, C.C.; Figlan, S.; Shimelis, H.A.; Motaung, T.E.; Sydenham, S.; Mwadzingeni, L.; Tsilo, T.J. Fusarium head blight of wheat: Pathogenesis and control strategies. Crop Prot. 2017, 91, 114–122. [Google Scholar] [CrossRef]
- Guenther, J.C.; Trail, F.J.M. The development and differentiation of Gibberella zeae (anamorph: Fusarium graminearum) during colonization of wheat. Mycologia 2005, 97, 229–237. [Google Scholar] [CrossRef]
- Mentges, M.; Glasenapp, A.; Boenisch, M.; Malz, S.; Henrissat, B.; Frandsen, R.J.N.; Güldener, U.; Münsterkötter, M.; Bormann, J.; Lebrun, M.H.; et al. Infection cushions of Fusarium graminearum are fungal arsenals for wheat infection. Mol. Plant Pathol. 2020, 21, 1070–1087. [Google Scholar] [CrossRef]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef]
- Ma, H.; Liu, Y.; Zhao, X.; Zhang, S.; Ma, H. Exploring and applying genes to enhance the resistance to Fusarium head blight in wheat. Front. Plant Sci. 2022, 13, 1026611. [Google Scholar] [CrossRef]
- Heitman, J.; Sun, S.; James, T.Y. Evolution of fungal sexual reproduction. Mycologia 2013, 105, 1–27. [Google Scholar] [CrossRef]
- Cavinder, B.; Sikhakolli, U.; Fellows, K.M.; Trail, F. Sexual development and ascospore discharge in Fusarium graminearum. J. Vis. Exp. 2012, 61, e3895. [Google Scholar] [CrossRef]
- Sun, M.; Bian, Z.; Luan, Q.; Chen, Y.; Wang, W.; Dong, Y.; Chen, L.; Hao, C.; Xu, J.R.; Liu, H. Stage-specific regulation of purine metabolism during infectious growth and sexual reproduction in Fusarium graminearum. New Phytol. 2021, 230, 757–773. [Google Scholar] [CrossRef]
- Zheng, Q.; Hou, R.; Zhang, J.; Ma, J.; Wu, Z.; Wang, G.; Wang, C.; Xu, J.R. The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PLoS ONE 2013, 8, e66980. [Google Scholar] [CrossRef]
- Kim, H.K.; Lee, T.; Yun, S.H. A putative pheromone signaling pathway is dispensable for self-fertility in the homothallic ascomycete Gibberella zeae. Fungal Genet. Biol. 2008, 45, 1188–1196. [Google Scholar] [CrossRef]
- Ding, M.; Cao, S.; Xu, D.; Xia, A.; Wang, Z.; Wang, W.; Duan, K.; Wu, C.; Wang, Q.; Liang, J.; et al. A non-pheromone GPCR is essential for meiosis and ascosporogenesis in the wheat scab fungus. Proc. Natl. Acad. Sci. USA 2023, 120, e2313034120. [Google Scholar] [CrossRef]
- Jiang, C.; Cao, S.; Wang, Z.; Xu, H.; Liang, J.; Liu, H.; Wang, G.; Ding, M.; Wang, Q.; Gong, C.; et al. An expanded subfamily of G-protein-coupled receptor genes in Fusarium graminearum required for wheat infection. Nat. Microbiol. 2019, 4, 1582–1591. [Google Scholar] [CrossRef]
- Yu, J.H. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 2006, 44, 145–154. [Google Scholar]
- Yu, H.Y.; Seo, J.A.; Kim, J.E.; Han, K.H.; Shim, W.B.; Yun, S.H.; Lee, Y.W. Functional analyses of heterotrimeric G protein G alpha and G beta subunits in Gibberella zeae. Microbiology 2008, 154, 392–401. [Google Scholar] [CrossRef]
- Chidiac, P.; Roy, A.A. Activity, regulation, and intracellular localization of RGS proteins. Recept. Channels 2003, 9, 135–147. [Google Scholar] [CrossRef]
- Park, A.R.; Cho, A.R.; Seo, J.A.; Min, K.; Son, H.; Lee, J.; Choi, G.J.; Kim, J.C.; Lee, Y.W. Functional analyses of regulators of G protein signaling in Gibberella zeae. Fungal Genet. Biol. 2012, 49, 511–520. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Jiang, C.; Xu, J.-R. Regulation of biotic interactions and responses to abiotic stresses by MAP kinase pathways in plant pathogenic fungi. Stress Biol. 2021, 1, 5. [Google Scholar] [CrossRef]
- Chen, A.; Liu, N.; Xu, C.; Wu, S.; Liu, C.; Qi, H.; Ren, Y.; Han, X.; Yang, K.; Liu, X.; et al. The STRIPAK complex orchestrates cell wall integrity signalling to govern the fungal development and virulence of Fusarium graminearum. Mol. Plant Pathol. 2023, 24, 1139–1153. [Google Scholar] [CrossRef]
- Urban, M.; Mott, E.; Farley, T.; Hammond-Kosack, K. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 2003, 4, 347–359. [Google Scholar] [CrossRef]
- Gu, Q.; Zhang, C.; Liu, X.; Ma, Z. A transcription factor FgSte12 is required for pathogenicity in Fusarium graminearum. Mol. Plant Pathol. 2015, 16, 12155. [Google Scholar] [CrossRef]
- Yang, C.; Liu, H.; Li, G.; Liu, M.; Yun, Y.; Wang, C.; Ma, Z.; Xu, J.R. The MADS-box transcription factor FgMcm1 regulates cell identity and fungal development in Fusarium graminearum. Environ. Microbiol. 2015, 17, 2762–2776. [Google Scholar] [CrossRef]
- Bluhm, B.H.; Zhao, X.; Flaherty, J.E.; Xu, J.R.; Dunkle, L.D. RAS2 regulates growth and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2007, 20, 627–636. [Google Scholar] [CrossRef]
- Zheng, D.; Zhang, S.; Zhou, X.; Wang, C.; Xiang, P.; Zheng, Q.; Xu, J.R. The FgHOG1 pathway regulates hyphal growth, stress responses, and plant infection in Fusarium graminearum. PLoS ONE 2012, 7, e49495. [Google Scholar] [CrossRef]
- Ren, J.; Zhang, Y.; Wang, Y.; Li, C.; Bian, Z.; Zhang, X.; Liu, H.; Xu, J.-R.; Jiang, C. Deletion of all three MAP kinase genes results in severe defects in stress responses and pathogenesis in Fusarium graminearum. Stress Biol. 2022, 2, 6. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, S.; Hou, R.; Zhao, Z.; Zheng, Q.; Xu, Q.; Zheng, D.; Wang, G.; Liu, H.; Gao, X.; et al. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 2011, 7, e1002460. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.; Wang, H.; Chen, L.; Zhang, J.; Sun, M.; Xu, J.R.; Wang, C. The PKR regulatory subunit of protein kinase A (PKA) is involved in the regulation of growth, sexual and asexual development, and pathogenesis in Fusarium graminearum. Mol. Plant Pathol. 2018, 19, 909–921. [Google Scholar] [CrossRef]
- Qin, J.; Wang, G.H.; Jiang, C.; Xu, J.R.; Wang, C.F. Fgk3 glycogen synthase kinase is important for development, pathogenesis, and stress responses in Fusarium graminearum. Sci. Rep. 2015, 5, 8504. [Google Scholar] [CrossRef]
- Lee, J.; Myong, K.; Kim, J.E.; Kim, H.K.; Yun, S.H.; Lee, Y.W. FgVelB globally regulates sexual reproduction, mycotoxin production and pathogenicity in the cereal pathogen Fusarium graminearum. Microbiology 2012, 158, 1723–1733. [Google Scholar] [CrossRef]
- Liu, K.; Wang, X.; Li, Y.; Shi, Y.; Ren, Y.; Wang, A.; Zhao, B.; Cheng, P.; Wang, B. Protein Disulfide Isomerase FgEps1 Is a Secreted Virulence Factor in Fusarium graminearum. J. Fungi 2023, 9, 1009. [Google Scholar] [CrossRef]
- Wu, C.; Chen, H.; Yuan, M.; Zhang, M.; Abubakar, Y.S.; Chen, X.; Zhong, H.; Zheng, W.; Zheng, H.; Zhou, J. FgAP1(σ) Is Critical for Vegetative Growth, Conidiation, Virulence, and DON Biosynthesis in Fusarium graminearum. J. Fungi 2023, 9, 145. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, H.H.; Park, J.; Kim, S.; Choi, S.; Moon, H.; Shin, J.; Kim, J.E.; Choi, G.J.; Seo, Y.S.; et al. Intron turnover is essential to the development and pathogenicity of the plant pathogenic fungus Fusarium graminearum. Commun. Biol. 2022, 5, 1129. [Google Scholar] [CrossRef]
- Han, X.; Li, Q.; Li, X.; Lv, X.; Zhang, L.; Zou, S.; Yu, J.; Dong, H.; Chen, L.; Liang, Y. Mitochondrial Porin Is Involved in Development, Virulence, and Autophagy in Fusarium graminearum. J. Fungi 2022, 8, 936. [Google Scholar] [CrossRef]
- Sun, F.; Lv, B.; Zhang, X.; Wang, C.; Zhang, L.; Chen, X.; Liang, Y.; Chen, L.; Zou, S.; Dong, H. The Endoplasmic Reticulum Cargo Receptor FgErv14 Regulates DON Production, Growth and Virulence in Fusarium graminearum. Life 2022, 12, 799. [Google Scholar] [CrossRef]
- Lou, Y.; Zhang, J.; Wang, G.; Fang, W.; Wang, S.; Abubakar, Y.S.; Zhou, J.; Wang, Z.; Zheng, W. Genome-Wide Characterization of PX Domain-Containing Proteins Involved in Membrane Trafficking-Dependent Growth and Pathogenicity of Fusarium graminearum. mBio 2021, 12, e0232421. [Google Scholar] [CrossRef]
- Wang, G.; Sun, P.; Sun, Z.; Zhu, J.; Yu, D.; Tang, Z.; Wang, Z.; Wang, C.; Zheng, H. Sgh1, an SR-like Protein, Is Involved in Fungal Development, Plant Infection, and Pre-mRNA Processing in Fusarium graminearum. J. Fungi 2022, 8, 1056. [Google Scholar] [CrossRef]
- Yuan, Y.; Mao, X.; Abubakar, Y.S.; Zheng, W.; Wang, Z.; Zhou, J.; Zheng, H. Genome-Wide Characterization of the RNA Exosome Complex in Relation to Growth, Development, and Pathogenicity of Fusarium graminearum. Microbiol. Spectr. 2023, 11, e05058-22. [Google Scholar] [CrossRef]
- Gong, C.; Huang, J.; Sun, D.; Xu, D.; Guo, Y.; Kang, J.; Niu, G.; Wang, C. FgSfl1 and Its Conserved PKA Phosphorylation Sites Are Important for Conidiation, Sexual Reproduction, and Pathogenesis in Fusarium graminearum. J. Fungi 2021, 7, 755. [Google Scholar] [CrossRef]
- Van Nguyen, T.; Kröger, C.; Bönnighausen, J.; Schäfer, W.; Bormann, J. The ATF/CREB transcription factor Atf1 is essential for full virulence, deoxynivalenol production, and stress tolerance in the cereal pathogen Fusarium graminearum. Mol. Plant Microbe Interact. 2013, 26, 1378–1394. [Google Scholar] [CrossRef]
- Rittenour, W.R.; Harris, S.D. Characterization of Fusarium graminearum Mes1 reveals roles in cell-surface organization and virulence. Fungal Genet. Biol. 2008, 45, 933–946. [Google Scholar] [CrossRef]
- Zhao, F.; Yuan, Z.; Wen, W.; Huang, Z.; Mao, X.; Zhou, M.; Hou, Y. FgMet3 and FgMet14 related to cysteine and methionine biosynthesis regulate vegetative growth, sexual reproduction, pathogenicity, and sensitivity to fungicides in Fusarium graminearum. Front. Plant Sci. 2022, 13, 1011709. [Google Scholar] [CrossRef]
- Tang, G.; Chen, A.; Dawood, D.H.; Liang, J.; Chen, Y.; Ma, Z. Capping proteins regulate fungal development, DON-toxisome formation and virulence in Fusarium graminearum. Mol. Plant Pathol. 2020, 21, 173–187. [Google Scholar] [CrossRef]
- Liu, N.; Fan, F.; Qiu, D.; Jiang, L. The transcription cofactor FgSwi6 plays a role in growth and development, carbendazim sensitivity, cellulose utilization, lithium tolerance, deoxynivalenol production and virulence in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 2013, 58, 42–52. [Google Scholar] [CrossRef]
- Jonkers, W.; Dong, Y.; Broz, K.; Corby Kistler, H. The Wor1-like Protein Fgp1 Regulates Pathogenicity, Toxin Synthesis and Reproduction in the Phytopathogenic Fungus Fusarium graminearum. PLoS Pathog. 2012, 8, e1002724. [Google Scholar] [CrossRef]
- Hu, S.; Zhou, X.; Gu, X.; Cao, S.; Wang, C.; Xu, J.R. The cAMP-PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2014, 27, 557–566. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, H.; Qi, L.; Zhang, S.; Zhou, X.; Zhang, Y.; Xu, J.R. FgKin1 kinase localizes to the septal pore and plays a role in hyphal growth, ascospore germination, pathogenesis, and localization of Tub1 beta-tubulins in Fusarium graminearum. New Phytol. 2014, 204, 943–954. [Google Scholar] [CrossRef]
- Chen, A.; Ren, Y.; Han, X.; Liu, C.; Zhou, Y.; Xu, C.; Qi, H.; Ma, Z.; Chen, Y. The COP9 signalosome complex regulates fungal development and virulence in the wheat scab fungus Fusarium graminearum. Front. Microbiol. 2023, 14, 1179676. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, Y.; Wang, W.; Cao, X.; Xu, H.; Liu, H.; Qi, J.; Jiang, C.; Wang, C. FgCsn12 is involved in the regulation of ascosporogenesis in the wheat scab fungus Fusarium graminearum. Int. J. Mol. Sci. 2022, 23, 10445. [Google Scholar] [CrossRef]
- Liang, J.; Fu, X.; Hao, C.; Bian, Z.; Liu, H.; Xu, J.R.; Wang, G. FgBUD14 is important for ascosporogenesis and involves both stage-specific alternative splicing and RNA editing during sexual reproduction. Environ. Microbiol. 2021, 23, 5052–5068. [Google Scholar] [CrossRef]
- Sun, S.; Wang, M.; Liu, C.; Tao, Y.; Wang, T.; Liang, Y.; Zhang, L.; Yu, J. FgLEU1 Is Involved in Leucine Biosynthesis, Sexual Reproduction, and Full Virulence in Fusarium graminearum. J. Fungi 2022, 8, 1090. [Google Scholar] [CrossRef]
- Trail, F.; Xu, H.; Loranger, R.; Gadoury, D. Physiological and environmental aspects of ascospore discharge in Gibberella zeae (anamorph Fusarium graminearum). Mycologia 2002, 94, 181–189. [Google Scholar] [CrossRef]
- David, R.F.; Reinisch, M.; Trail, F.; Marr, L.C.; Schmale, D.G. Compression tests of Fusarium graminearum ascocarps provide insights into the strength of the perithecial wall and the quantity of ascospores. Fungal Genet. Biol. 2016, 96, 25–32. [Google Scholar] [CrossRef]
- Min, K.; Lee, J.; Kim, J.C.; Kim, S.G.; Kim, Y.H.; Vogel, S.; Trail, F.; Lee, Y.W. A novel gene, ROA, is required for normal morphogenesis and discharge of ascospores in Gibberella zeae. Eukaryot. Cell 2010, 9, 1495–1503. [Google Scholar] [CrossRef]
- Hallen, H.E.; Trail, F. The L-type calcium ion channel cch1 affects ascospore discharge and mycelial growth in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot. Cell 2008, 7, 415–424. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, X.; Sun, M.; Liu, H.; Xu, J.R. The FgSRP1 SR-protein gene is important for plant infection and pre-mRNA processing in Fusarium graminearum. Environ. Microbiol. 2017, 19, 4065–4079. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, W.; Cheng, J.; Xie, J.; Fu, Y.; Jiang, D.; Lin, Y. lncRsp1, a long noncoding RNA, influences Fgsp1 expression and sexual reproduction in Fusarium graminearum. Mol. Plant Pathol. 2022, 23, 265–277. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, S.; Zhang, Q.; Tao, Y.; Wang, C.; Xu, J.R. FgSKN7 and FgATF1 have overlapping functions in ascosporogenesis, pathogenesis and stress responses in Fusarium graminearum. Environ. Microbiol. 2015, 17, 1245–1260. [Google Scholar] [CrossRef]
- Son, H.; Lee, J.; Lee, Y.W. A novel gene, GEA1, is required for ascus cell-wall development in the ascomycete fungus Fusarium graminearum. Microbiology 2013, 159, 1077–1085. [Google Scholar] [CrossRef]
- Sikhakolli, U.R.; López-Giráldez, F.; Li, N.; Common, R.; Townsend, J.P.; Trail, F. Transcriptome analyses during fruiting body formation in Fusarium graminearum and Fusarium verticillioides reflect species life history and ecology. Fungal Genet. Biol. 2012, 49, 663–673. [Google Scholar] [CrossRef]
- Ni, M.; Feretzaki, M.; Sun, S.; Wang, X.; Heitman, J. Sex in fungi. Annu. Rev. Genet. 2011, 45, 405–430. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Q.; He, Y.; Chen, L.; Hao, C.; Jiang, C.; Li, Y.; Dai, Y.; Kang, Z.; Xu, J.R. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016, 26, 499–509. [Google Scholar] [CrossRef]
- Wang, X.; Hsueh, Y.P.; Li, W.; Floyd, A.; Skalsky, R.; Heitman, J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010, 24, 2566–2582. [Google Scholar] [CrossRef]
- Cuomo, C.A.; Güldener, U.; Xu, J.-R.; Trail, F.; Turgeon, B.G.; Di Pietro, A.; Walton, J.D.; Ma, L.-J.; Baker, S.E.; Rep, M.; et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 2007, 317, 1400–1402. [Google Scholar] [CrossRef]
- Cambareri, E.B.; Jensen, B.C.; Schabtach, E.; Selker, E.U. Repeat-induced G-C to A-T mutations in Neurospora. Science 1989, 244, 1571–1575. [Google Scholar] [CrossRef]
- Selker, E.U.; Tountas, N.A.; Cross, S.H.; Margolin, B.S.; Murphy, J.G.; Bird, A.P.; Freitag, M.J.N. The methylated component of the Neurospora crassa genome. Nature 2003, 422, 893–897. [Google Scholar] [CrossRef]
- Shiu, P.K.; Raju, N.B.; Zickler, D.; Metzenberg, R.L. Meiotic silencing by unpaired DNA. Cell 2001, 107, 905–916. [Google Scholar] [CrossRef]
- Son, H.; Min, K.; Lee, J.; Raju, N.B.; Lee, Y.W. Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol. 2011, 115, 1290–1302. [Google Scholar] [CrossRef]
- Bian, Z.; Ni, Y.; Xu, J.R.; Liu, H. A-to-I mRNA editing in fungi: Occurrence, function, and evolution. Cell. Mol. Life Sci. 2019, 76, 329–340. [Google Scholar] [CrossRef]
- Hao, C.; Yin, J.; Sun, M.; Wang, Q.; Liang, J.; Bian, Z.; Liu, H.; Xu, J.R. The meiosis-specific APC activator FgAMA1 is dispensable for meiosis but important for ascosporogenesis in Fusarium graminearum. Mol. Microbiol. 2019, 111, 1245–1262. [Google Scholar] [CrossRef]
- Cao, S.; He, Y.; Hao, C.; Xu, Y.; Zhang, H.; Wang, C.; Liu, H.; Xu, J.R. RNA editing of the AMD1 gene is important for ascus maturation and ascospore discharge in Fusarium graminearum. Sci.Rep. 2017, 7, 4617. [Google Scholar] [CrossRef]
- Feng, C.; Cao, X.; Du, Y.; Chen, Y.; Xin, K.; Zou, J.; Jin, Q.; Xu, J.R.; Liu, H. Uncovering cis-regulatory elements important for A-to-I RNA editing in Fusarium graminearum. mBio 2022, 13, e0187222. [Google Scholar] [CrossRef]
- Xin, K.; Zhang, Y.; Fan, L.; Qi, Z.; Feng, C.; Wang, Q.; Jiang, C.; Xu, J.R.; Liu, H. Experimental evidence for the functional importance and adaptive advantage of A-to-I RNA editing in fungi. Proc. Natl. Acad. Sci. USA 2023, 120, e2219029120. [Google Scholar] [CrossRef]
- Qi, Z.; Lu, P.; Long, X.; Cao, X.; Wu, M.; Xin, K.; Xue, T.; Gao, X.; Huang, Y.; Wang, Q.; et al. Adaptive advantages of restorative RNA editing in fungi for resolving survival-reproduction trade-offs. Sci. Adv. 2024, 10, eadk6130. [Google Scholar] [CrossRef]
- Son, H.; Park, A.R.; Lim, J.Y.; Shin, C.; Lee, Y.W. Genome-wide exonic small interference RNA-mediated gene silencing regulates sexual reproduction in the homothallic fungus Fusarium graminearum. PLoS Genet. 2017, 13, e1006595. [Google Scholar] [CrossRef]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Chang, S.S.; Zhang, Z.; Liu, Y. RNA interference pathways in fungi: Mechanisms and functions. Annu. Rev. Microbiol. 2012, 66, 305–323. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Ruiz-Vázquez, R.M. Functional diversity of RNAi-associated sRNAs in fungi. Int. J. Mol. Sci. 2013, 14, 15348–15360. [Google Scholar] [CrossRef]
- Nguyen, Q.; Iritani, A.; Ohkita, S.; Vu, B.V.; Yokoya, K.; Matsubara, A.; Ikeda, K.-i.; Suzuki, N.; Nakayashiki, H. A fungal Argonaute interferes with RNA interference. Nucleic Acids Res. 2018, 46, 2495–2508. [Google Scholar] [CrossRef]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef]
- Brodersen, P.; Voinnet, O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006, 22, 268–280. [Google Scholar] [CrossRef]
- Kim, H.K.; Jo, S.M.; Kim, G.Y.; Kim, D.W.; Kim, Y.K.; Yun, S.H. A Large-Scale Functional Analysis of Putative Target Genes of Mating-Type Loci Provides Insight into the Regulation of Sexual Development of the Cereal Pathogen Fusarium graminearum. PLoS Genet. 2015, 11, e1005486. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, J.; Wang, Y.; Lin, J.; Fu, Y.; Xie, J.; Jiang, D.; Chen, T.; Liu, H.; Cheng, J. Dicer-Like Proteins Regulate Sexual Development via the Biogenesis of Perithecium-Specific MicroRNAs in a Plant Pathogenic Fungus Fusarium graminearum. Front. Microbiol. 2018, 9, 818. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef]
- Brown, N.A.; Evans, J.; Mead, A.; Hammond-Kosack, K.E. A spatial temporal analysis of the Fusarium graminearum transcriptome during symptomless and symptomatic wheat infection. Mol. Plant Pathol. 2017, 18, 1295–1312. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, B.; Zheng, W.; Wang, L.; Cai, X.; Yang, J.; Song, R.; Yang, S.; Wang, Y.; Xiao, J.; et al. Recognition of glycoside hydrolase 12 proteins by the immune receptor RXEG1 confers Fusarium head blight resistance in wheat. Plant Biotechnol. J. 2023, 21, 769–781. [Google Scholar] [CrossRef]
- Carere, J.; Benfield, A.H.; Ollivier, M.; Liu, C.J.; Kazan, K.; Gardiner, D.M. A tomatinase-like enzyme acts as a virulence factor in the wheat pathogen Fusarium graminearum. Fungal Genet. Biol. 2017, 100, 33–41. [Google Scholar] [CrossRef]
- Hao, G.; McCormick, S.; Vaughan, M.M.; Naumann, T.A.; Kim, H.S.; Proctor, R.; Kelly, A.; Ward, T.J. Fusarium graminearum arabinanase (Arb93B) enhances Wheat Head Blight susceptibility by suppressing plant immunity. Mol. Plant Microbe Interact. 2019, 32, 888–898. [Google Scholar] [CrossRef]
- Guo, Y.; Yao, S.; Yuan, T.; Wang, Y.; Zhang, D.; Tang, W. The spatiotemporal control of KatG2 catalase-peroxidase contributes to the invasiveness of Fusarium graminearum in host plants. Mol. Plant Pathol. 2019, 20, 685–700. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Y.; Tian, M.; Dai, K.; Zheng, W.; Liu, Z.; Yang, S.; Liu, X.; Shi, D.; Zhang, H.; et al. Fg12 ribonuclease secretion contributes to Fusarium graminearum virulence and induces plant cell death. J. Integr. Plant Biol. 2021, 63, 365–377. [Google Scholar] [CrossRef]
- Cantu, D.; Vicente, A.R.; Labavitch, J.M.; Bennett, A.B.; Powell, A.L. Strangers in the matrix: Plant cell walls and pathogen susceptibility. Trends Plant Sci. 2008, 13, 610–617. [Google Scholar] [CrossRef]
- Blümke, A.; Falter, C.; Herrfurth, C.; Sode, B.; Bode, R.; Schäfer, W.; Feussner, I.; Voigt, C.A. Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol. 2014, 165, 346–358. [Google Scholar] [CrossRef]
- Brown, N.A.; Antoniw, J.; Hammond-Kosack, K.E. The predicted secretome of the plant pathogenic fungus Fusarium graminearum: A refined comparative analysis. PLoS ONE 2012, 7, e33731. [Google Scholar] [CrossRef]
- Lu, S.; Edwards, M.C. Genome-wide analysis of small secreted cysteine-rich proteins identifies candidate effector proteins potentially involved in Fusarium graminearum-Wheat interactions. Phytopathology 2016, 106, 166–176. [Google Scholar] [CrossRef]
- Rocher, F.; Alouane, T.; Philippe, G.; Martin, M.L.; Label, P.; Langin, T.; Bonhomme, L. Fusarium graminearum Infection Strategy in Wheat Involves a Highly Conserved Genetic Program That Controls the Expression of a Core Effectome. Int. J. Mol. Sci. 2022, 23, 1914. [Google Scholar] [CrossRef]
- Jiang, C.; Hei, R.; Yang, Y.; Zhang, S.; Wang, Q.; Wang, W.; Zhang, Q.; Yan, M.; Zhu, G.; Huang, P.; et al. An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 2020, 11, 4382. [Google Scholar] [CrossRef]
- Xu, Q.; Hu, S.; Jin, M.; Xu, Y.; Jiang, Q.; Ma, J.; Zhang, Y.; Qi, P.; Chen, G.; Jiang, Y.; et al. The N-terminus of a Fusarium graminearum-secreted protein enhances broad-spectrum disease resistance in plants. Mol. Plant Pathol. 2022, 23, 1751–1764. [Google Scholar] [CrossRef]
- Zuo, N.; Bai, W.-Z.; Wei, W.-Q.; Yuan, T.-L.; Zhang, D.; Wang, Y.-Z.; Tang, W.-H. Fungal CFEM effectors negatively regulate a maize wall-associated kinase by interacting with its alternatively spliced variant to dampen resistance. Cell Rep. 2022, 41, 111877. [Google Scholar] [CrossRef]
- Lee, H.J.; Ryu, D. Worldwide Occurrence of Mycotoxins in Cereals and Cereal-Derived Food Products: Public Health Perspectives of Their Co-occurrence. J. Agric. Food Chem. 2017, 65, 7034–7051. [Google Scholar] [CrossRef]
- Pestka, J.J. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 2010, 84, 663–679. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; O’Donnell, K.; Ward, T.J.; Fujimura, M.; Hamamoto, H.; Shibata, T.; Yamaguchi, I. The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett. 2003, 539, 105–110. [Google Scholar] [CrossRef]
- Ilgen, P.; Hadeler, B.; Maier, F.J.; Schafer, W. Developing kernel and rachis node induce the trichothecene pathway of Fusarium graminearum during wheat head infection. Mol. Plant Microbe Interact. 2009, 22, 899–908. [Google Scholar] [CrossRef]
- Maier, F.J.; Miedaner, T.; Hadeler, B.; Felk, A.; Salomon, S.; Lemmens, M.; Kassner, H.; Schafer, W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 2006, 7, 449–461. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Seong, K.Y.; Pasquali, M.; Zhou, X.; Song, J.; Hilburn, K.; McCormick, S.; Dong, Y.; Xu, J.R.; Kistler, H.C. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 2009, 72, 354–367. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, C.; Wu, C.; Sun, P.; Hou, R.; Liu, H.; Wang, C.; Xu, J.R. TRI6 and TRI10 play different roles in the regulation of deoxynivalenol (DON) production by cAMP signalling in Fusarium graminearum. Environ. Microbiol. 2016, 18, 3689–3701. [Google Scholar] [CrossRef]
- Huang, P.; Yu, X.; Liu, H.; Ding, M.; Wang, Z.; Xu, J.-R.; Jiang, C. Regulation of TRI5 expression and deoxynivalenol biosynthesis by a long non-coding RNA in Fusarium graminearum. Nat. Commun. 2024, 15, 1216. [Google Scholar] [CrossRef]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef]
- Yu, F.; Gu, Q.; Yun, Y.; Yin, Y.; Xu, J.R.; Shim, W.B.; Ma, Z. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014, 203, 219–232. [Google Scholar] [CrossRef]
- Yin, T.; Zhang, Q.; Wang, J.; Liu, H.; Wang, C.; Xu, J.R.; Jiang, C. The cyclase-associated protein FgCap1 has both protein kinase A-dependent and -independent functions during deoxynivalenol production and plant infection in Fusarium graminearum. Mol. Plant Pathol. 2018, 19, 552–563. [Google Scholar] [CrossRef]
- Hou, Z.; Xue, C.; Peng, Y.; Katan, T.; Kistler, H.C.; Xu, J.R. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 2002, 15, 1119–1127. [Google Scholar] [CrossRef]
- Yun, Y.; Liu, Z.; Zhang, J.; Shim, W.B.; Chen, Y.; Ma, Z. The MAPKK FgMkk1 of Fusarium graminearum regulates vegetative differentiation, multiple stress response, and virulence via the cell wall integrity and high-osmolarity glycerol signaling pathways. Environ. Microbiol. 2014, 16, 2023–2037. [Google Scholar] [CrossRef]
- Gu, Q.; Chen, Y.; Liu, Y.; Zhang, C.; Ma, Z. The transmembrane protein FgSho1 regulates fungal development and pathogenicity via the MAPK module Ste50-Ste11-Ste7 in Fusarium graminearum. New Phytol. 2015, 206, 315–328. [Google Scholar] [CrossRef]
- Jenczmionka, N.J.; Maier, F.J.; Losch, A.P.; Schafer, W. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 2003, 43, 87–95. [Google Scholar] [CrossRef]
- Jiang, J.; Yun, Y.; Fu, J.; Shim, W.B.; Ma, Z. Involvement of a putative response regulator FgRrg-1 in osmotic stress response, fungicide resistance and virulence in Fusarium graminearum. Mol. Plant Pathol. 2011, 12, 425–436. [Google Scholar] [CrossRef]
- Van Thuat, N.; Schäfer, W.; Bormann, J. The stress-activated protein kinase FgOS-2 is a key regulator in the life cycle of the cereal pathogen Fusarium graminearum. Mol. Plant Microbe Interact. 2012, 25, 1142–1156. [Google Scholar] [CrossRef]
- Ochiai, N.; Tokai, T.; Nishiuchi, T.; Takahashi-Ando, N.; Fujimura, M.; Kimura, M. Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum. Biochem. Biophys. Res. Commun. 2007, 363, 639–644. [Google Scholar] [CrossRef]
- Xu, C.; Wang, J.; Zhang, Y.; Luo, Y.; Zhao, Y.; Chen, Y.; Ma, Z. The transcription factor FgStuA regulates virulence and mycotoxin biosynthesis via recruiting the SAGA complex in Fusarium graminearum. New Phytol. 2023, 240, 2455–2467. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, S.; Ju, Z.; Zhang, C.; Tang, G.; Wang, J.; Wen, Z.; Chen, W.; Ma, Z. Contribution of peroxisomal docking machinery to mycotoxin biosynthesis, pathogenicity and pexophagy in the plant pathogenic fungus Fusarium graminearum. Environ. Microbiol. 2018, 20, 3224–3245. [Google Scholar] [CrossRef]
- Xu, L.; Wang, H.; Zhang, C.; Wang, J.; Chen, A.; Chen, Y.; Ma, Z. System-wide characterization of subtilases reveals that subtilisin-like protease FgPrb1 of Fusarium graminearum regulates fungal development and virulence. Fungal Genet. Biol. 2020, 144, 103449. [Google Scholar] [CrossRef]
- Reyes-Dominguez, Y.; Boedi, S.; Sulyok, M.; Wiesenberger, G.; Stoppacher, N.; Krska, R.; Strauss, J. Heterochromatin influences the secondary metabolite profile in the plant pathogen Fusarium graminearum. Fungal Genet. Biol. 2012, 49, 39–47. [Google Scholar] [CrossRef]
- Connolly, L.R.; Smith, K.M.; Freitag, M. The Fusarium graminearum Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters. PLoS Genet. 2013, 9, e1003916. [Google Scholar] [CrossRef]
- Xu, H.; Ye, M.; Xia, A.; Jiang, H.; Huang, P.; Liu, H.; Hou, R.; Wang, Q.; Li, D.; Xu, J.R.; et al. The Fng3 ING protein regulates H3 acetylation and H4 deacetylation by interacting with two distinct histone-modifying complexes. New Phytol. 2022, 235, 2350–2364. [Google Scholar] [CrossRef]
- Jiang, H.; Xia, A.; Ye, M.; Ren, J.; Li, D.; Liu, H.; Wang, Q.; Lu, P.; Wu, C.; Xu, J.R.; et al. Opposing functions of Fng1 and the Rpd3 HDAC complex in H4 acetylation in Fusarium graminearum. PLoS Genet. 2020, 16, e1009185. [Google Scholar] [CrossRef]
- Boenisch, M.J.; Broz, K.L.; Purvine, S.O.; Chrisler, W.B.; Nicora, C.D.; Connolly, L.R.; Freitag, M.; Baker, S.E.; Kistler, H.C. Structural reorganization of the fungal endoplasmic reticulum upon induction of mycotoxin biosynthesis. Sci. Rep. 2017, 7, 44296. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Wang, J.; Chen, Y.; Hou, T.; Zhao, Y.; Ma, Z. The sphinganine C4-hydroxylase FgSur2 regulates sensitivity to azole antifungal agents and virulence of Fusarium graminearum. Microbiol. Res. 2023, 271, 127347. [Google Scholar] [CrossRef]
- Chen, A.; Ju, Z.; Wang, J.; Wang, J.; Wang, H.; Wu, J.; Yin, Y.; Zhao, Y.; Ma, Z.; Chen, Y. The RasGEF FgCdc25 regulates fungal development and virulence in Fusarium graminearum via cAMP and MAPK signalling pathways. Environ. Microbiol. 2020, 22, 5109–5124. [Google Scholar] [CrossRef]
- Tang, G.; Chen, Y.; Xu, J.R.; Kistler, H.C.; Ma, Z. The fungal myosin I is essential for Fusarium toxisome formation. PLoS Pathog. 2018, 14, e1006827. [Google Scholar] [CrossRef]
- Zhou, Z.; Duan, Y.; Zhang, J.; Lu, F.; Zhu, Y.; Shim, W.B.; Zhou, M. Microtubule-assisted mechanism for toxisome assembly in Fusarium graminearum. Mol. Plant Pathol. 2021, 22, 163–174. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, X.; Yin, Y.; Ma, Z. Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum. PLoS ONE 2011, 6, e28291. [Google Scholar] [CrossRef]
- Merhej, J.; Richard-Forget, F.; Barreau, C. Regulation of trichothecene biosynthesis in Fusarium: Recent advances and new insights. Appl. Microbiol. Biotechnol. 2011, 91, 519–528. [Google Scholar] [CrossRef]
- Jiao, F.; Kawakami, A.; Nakajima, T. Effects of different carbon sources on trichothecene production and Tri gene expression by Fusarium graminearum in liquid culture. FEMS Microbiol. Lett. 2008, 285, 212–219. [Google Scholar] [CrossRef]
- Zhang, H.; Wolf-Hall, C. The effect of different carbon sources on phenotypic expression by Fusarium graminearum strains. Eur. J. Plant Pathol. 2010, 127, 137–148. [Google Scholar] [CrossRef]
- Kovács, B.; Kovács, A.; Pál, M.; Spitkó, T.; Marton, C.L.; Szőke, C. Changes in polyamine contents during Fusarium graminearum and Fusarium verticillioides inoculation in maize seedlings with or without seed-priming. Biol. Futura 2023, 74, 145–157. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Kazan, K.; Praud, S.; Torney, F.J.; Rusu, A.; Manners, J.M. Early activation of wheat polyamine biosynthesis during Fusarium head blight implicates putrescine as an inducer of trichothecene mycotoxin production. BMC Plant Biol. 2010, 10, 289. [Google Scholar] [CrossRef]
- Tang, G.F.; Xia, H.X.; Liang, J.T.; Ma, Z.H.; Liu, W.D. Spermidine Is Critical for Growth, Development, Environmental Adaptation, and Virulence in Fusarium graminearum. Front. Microbiol. 2021, 12, 765398. [Google Scholar] [CrossRef]
- Ma, T.; Zhang, L.; Wang, M.; Li, Y.; Jian, Y.; Wu, L.; Kistler, H.C.; Ma, Z.; Yin, Y. Plant defense compound triggers mycotoxin synthesis by regulating H2B ub1 and H3K4 me2/3 deposition. New Phytol. 2021, 232, 2106–2123. [Google Scholar] [CrossRef]
- Hou, R.; Jiang, C.; Zheng, Q.; Wang, C.; Xu, J.R. The AreA transcription factor mediates the regulation of deoxynivalenol (DON) synthesis by ammonium and cyclic adenosine monophosphate (cAMP) signalling in Fusarium graminearum. Mol. Plant Pathol. 2015, 16, 987–999. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Osborne, S.; Kazan, K.; Manners, J.M.J.M. Low pH regulates the production of deoxynivalenol by Fusarium graminearum. Microbiology 2009, 155, 3149–3156. [Google Scholar] [CrossRef]
- Merhej, J.; Boutigny, A.-L.; Pinson-Gadais, L.; Richard-Forget, F.; Barreau, C.J.F.A. Acidic pH as a determinant of TRI gene expression and trichothecene B biosynthesis in Fusarium graminearum. Food Addit. Contam. 2010, 27, 710–717. [Google Scholar] [CrossRef]
- Merhej, J.; Richard-Forget, F.; Barreau, C. The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet. Biol. 2011, 48, 275–284. [Google Scholar] [CrossRef]
- Gu, Q.; Wang, Y.; Zhao, X.; Yuan, B.; Zhang, M.; Tan, Z.; Zhang, X.; Chen, Y.; Wu, H.; Luo, Y.; et al. Inhibition of histone acetyltransferase GCN5 by a transcription factor FgPacC controls fungal adaption to host-derived iron stress. Nucleic Acids Res. 2022, 50, 6190–6210. [Google Scholar] [CrossRef]
- Audenaert, K.; Callewaert, E.; Höfte, M.; De Saeger, S.; Haesaert, G. Hydrogen peroxide induced by the fungicide prothioconazole triggers deoxynivalenol (DON) production by Fusarium graminearum. BMC Microbiol. 2010, 10, 112. [Google Scholar] [CrossRef]
- Luo, K.; Guo, J.; He, D.; Li, G.; Ouellet, T. Deoxynivalenol accumulation and detoxification in cereals and its potential role in wheat–Fusarium graminearum interactions. aBIOTECH 2023, 4, 155–171. [Google Scholar] [CrossRef]
- Luo, K.; Ouellet, T.; Zhao, H.; Wang, X.; Kang, Z. Wheat-Fusarium graminearum Interactions Under Sitobion avenae Influence: From Nutrients and Hormone Signals. Front. Nutr. 2021, 8, 703293. [Google Scholar] [CrossRef]
- Schlemmer, T.; Lischka, R.; Wegner, L.; Ehlers, K.; Biedenkopf, D.; Koch, A. Extracellular vesicles isolated from dsRNA-sprayed barley plants exhibit no growth inhibition or gene silencing in Fusarium graminearum. Fungal Biol. Biotechnol. 2022, 9, 14. [Google Scholar] [CrossRef]
- Wang, M.; Wu, L.; Mei, Y.; Zhao, Y.; Ma, Z.; Zhang, X.; Chen, Y. Host-induced gene silencing of multiple genes of Fusarium graminearum enhances resistance to Fusarium head blight in wheat. Plant Biotechnol. J. 2020, 18, 2373–2375. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Ruan, S.; Nzabanita, C.; Wang, Y.; Guo, L. A Mycovirus VIGS Vector Confers Hypovirulence to a Plant Pathogenic Fungus to Control Wheat FHB. Adv. Sci. 2023, 10, e2302606. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, X.; Liu, W. Targeting wheat fusarium head blight with mycovirus-mediated VIGS. Trends Microbiol. 2023, 31, 1197–1198. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, G.; Yang, Q.; Liao, Y.; Sun, D.; Tang, Z.; Wang, G.; Xu, M.; Wang, C.; Kang, J. Advances in Understanding Fusarium graminearum: Genes Involved in the Regulation of Sexual Development, Pathogenesis, and Deoxynivalenol Biosynthesis. Genes 2024, 15, 475. https://doi.org/10.3390/genes15040475
Niu G, Yang Q, Liao Y, Sun D, Tang Z, Wang G, Xu M, Wang C, Kang J. Advances in Understanding Fusarium graminearum: Genes Involved in the Regulation of Sexual Development, Pathogenesis, and Deoxynivalenol Biosynthesis. Genes. 2024; 15(4):475. https://doi.org/10.3390/genes15040475
Chicago/Turabian StyleNiu, Gang, Qing Yang, Yihui Liao, Daiyuan Sun, Zhe Tang, Guanghui Wang, Ming Xu, Chenfang Wang, and Jiangang Kang. 2024. "Advances in Understanding Fusarium graminearum: Genes Involved in the Regulation of Sexual Development, Pathogenesis, and Deoxynivalenol Biosynthesis" Genes 15, no. 4: 475. https://doi.org/10.3390/genes15040475
APA StyleNiu, G., Yang, Q., Liao, Y., Sun, D., Tang, Z., Wang, G., Xu, M., Wang, C., & Kang, J. (2024). Advances in Understanding Fusarium graminearum: Genes Involved in the Regulation of Sexual Development, Pathogenesis, and Deoxynivalenol Biosynthesis. Genes, 15(4), 475. https://doi.org/10.3390/genes15040475






