Genome and Epigenome Disorders and Male Infertility: Feedback from 15 Years of Clinical and Research Experience
Abstract
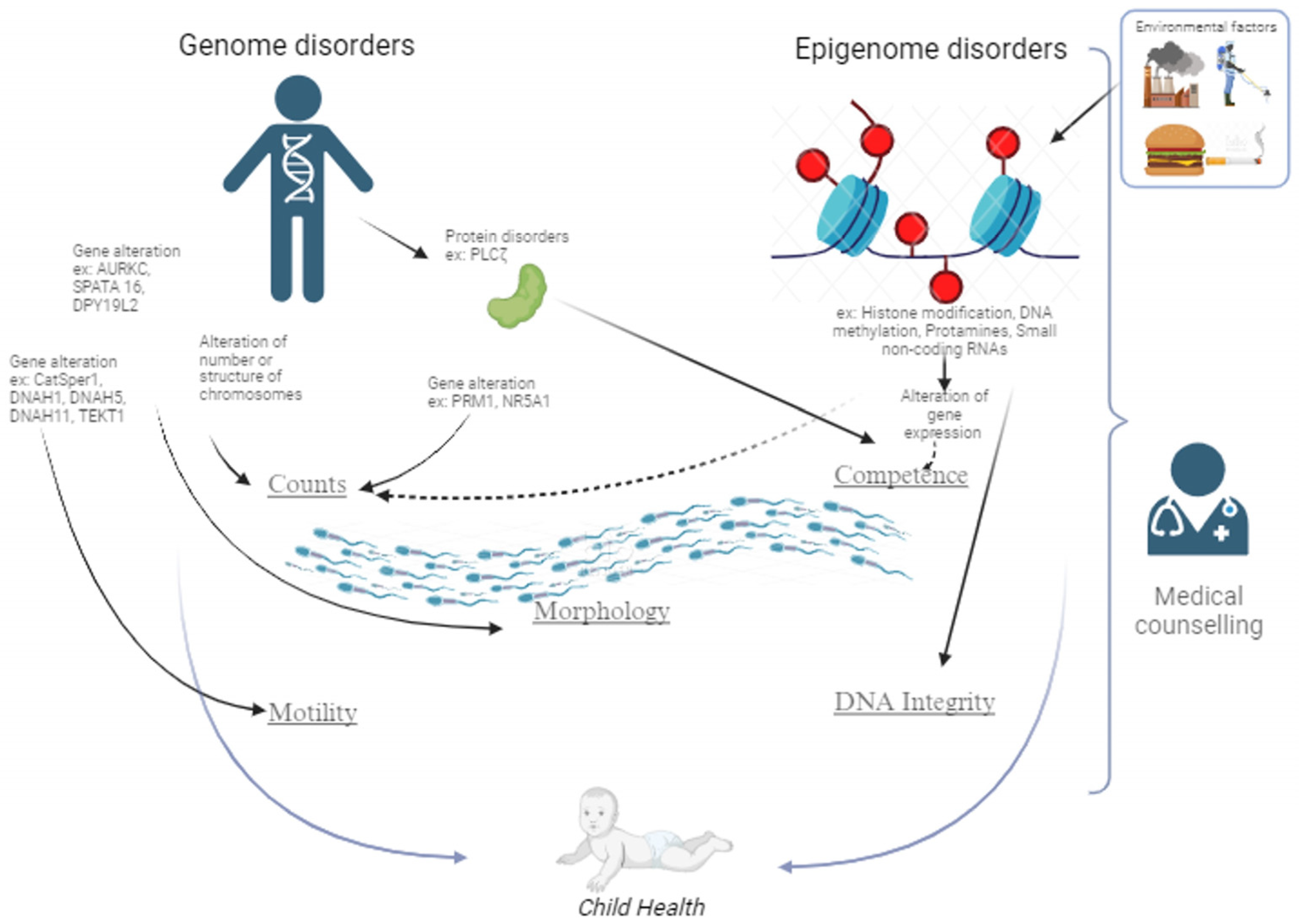
1. Introduction
2. Genetics and Male Infertility
3. Sperm Genome Decays
3.1. DNA Fragmentation
3.2. Sperm Chromatin Decondensation
3.3. Sperm Parameters Declining and Specific Genes Defects
3.3.1. Reduced Sperm Counts
3.3.2. Asthenozoospermia
3.3.3. Teratozoospermia
3.3.4. Protein Dysregulation
3.3.5. Epigenetic Marks
3.3.5.1. Histones
3.3.5.2. Protamines
3.3.5.3. RNA Associated Gene Silencing
3.3.6. Methylome Unbalance
3.3.7. Nuclear Architecture Disorganization
3.3.8. Peripheral Free Circulating DNA and Sperm Parameters
4. What for the Future
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boitrelle, F.; Shah, R.; Saleh, R.; Henkel, R.; Kandil, H.; Chung, E.; Vogiatzi, P.; Zini, A.; Arafa, M.; Agarwal, A. The Sixth Edition of the WHO Manual for Human Semen Analysis: A Critical Review and SWOT Analysis. Life 2021, 11, 1368. [Google Scholar] [CrossRef]
- Laan, M.; Kasak, L.; Punab, M. Translational aspects of novel findings in genetics of male infertility-status quo 2021. Br. Med. Bull. 2021, 140, 5–22. [Google Scholar] [CrossRef]
- Tiepolo, L.; Zuffardi, O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum. Genet. 1976, 34, 119–124. [Google Scholar] [CrossRef]
- Luddi, A.; Margollicci, M.; Gambera, L.; Serafini, F.; Cioni, M.; De Leo, V.; Balestri, P.; Piomboni, P. Spermatogenesis in a man with complete deletion of USP9Y. N. Engl. J. Med. 2009, 360, 881–885. [Google Scholar] [CrossRef]
- Ditton, H.J.; Zimmer, J.; Kamp, C.; Rajpert-De Meyts, E.; Vogt, P.H. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 2004, 13, 2333–2341. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, H.; Sabbaghian, M.; Vogiatzi, P.; Rambhatla, A.; Agarwal, A.; Colpi, G.M.; Gilani, M.A.S. Bridging the Gap between AZF Microdeletions and Karyotype: Twelve Years’ Experience of an Infertility Center. World J. Men’s Health 2023, 41, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Jamsai, D.; O’Bryan, M.K. Mouse models in male fertility research. Asian J. Androl. 2011, 13, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Houston, B.J.; Riera-Escamilla, A.; Wyrwoll, M.J.; Salas-Huetos, A.; Xavier, M.J.; Nagirnaja, L.; Friedrich, C.; Conrad, D.F.; I Aston, K.; Krausz, C.; et al. A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene-disease relationships. Hum. Reprod. Update 2021, 28, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, D.V.S.; Shah, R.; Gajbhiye, R.K. Genetics of Male Infertility—Present and Future: A Narrative Review. J. Hum. Reprod. Sci. 2021, 14, 217–227. [Google Scholar] [PubMed]
- Cho, C.-L.; Agarwal, A. Role of sperm DNA fragmentation in male factor infertility: A systematic review. Arab. J. Urol. 2018, 16, 21–34. [Google Scholar] [CrossRef]
- Gonzalez, D.C.; Ory, J.; Blachman-Braun, R.; Nackeeran, S.; Best, J.C.; Ramasamy, R. Advanced Paternal Age and Sperm DNA Fragmentation: A Systematic Review. World J. Men’s Health 2022, 40, 104–115. [Google Scholar] [CrossRef]
- Belloc, S.; Hazout, A.; Zini, A.; Merviel, P.; Cabry, R.; Chahine, H.; Copin, H.; Benkhalifa, M. How to overcome male infertility after 40: Influence of paternal age on fertility. Maturitas 2014, 78, 22–29. [Google Scholar] [CrossRef]
- Benkhalifa, M.; Montjean, D.; Belloc, S.; Dalleac, A.; Ducasse, M.; Boyer, P.; Merviel, P.; Copin, H. Emerging molecular methods for male infertility investigation. Expert Rev. Mol. Diagn. 2014, 14, 37–45. [Google Scholar] [CrossRef]
- Liebaers, I.; Bonduelle, M.; Van Assche, E.; Devroey, P.; Van Steirteghem, A. Sex chromosome abnormalities after intracytoplasmic sperm injection. Lancet 1995, 346, 1095. [Google Scholar] [CrossRef]
- Fernández-Gonzalez, R.; Moreira, P.N.; Pérez-Crespo, M.; Sánchez-Martín, M.; Ramirez, M.A.; Pericuesta, E.; Bilbao, A.; Bermejo-Alvarez, P.; Hourcade, J.d.D.; de Fonseca, F.R.; et al. Long-term effects of mouse intracytoplasmic sperm injection with DNA-fragmented sperm on health and behavior of adult offspring. Biol. Reprod. 2008, 78, 761–772. [Google Scholar] [CrossRef]
- Zini, A. Are sperm chromatin and DNA defects relevant in the clinic? Syst. Biol. Reprod. Med. 2011, 57, 78–85. [Google Scholar] [CrossRef]
- Robinson, L.; Gallos, I.D.; Conner, S.J.; Rajkhowa, M.; Miller, D.; Lewis, S.; Kirkman-Brown, J.; Coomarasamy, A. The effect of sperm DNA fragmentation on miscarriage rates: A systematic review and meta-analysis. Hum. Reprod. 2012, 27, 2908–2917. [Google Scholar] [CrossRef] [PubMed]
- Benchaib, M.; Ajina, M.; Lornage, J.; Niveleau, A.; Durand, P.; Guérin, J.F. Quantitation by image analysis of global DNA methylation in human spermatozoa and its prognostic value in in vitro fertilization: A preliminary study. Fertil. Steril. 2003, 80, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Benchaib, M.; Lornage, J.; Mazoyer, C.; Lejeune, H.; Salle, B.; François Guerin, J. Sperm deoxyribonucleic acid fragmentation as a prognostic indicator of assisted reproductive technology outcome. Fertil. Steril. 2007, 87, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Palermo, G.D.; Neri, Q.V.; Cozzubbo, T.; Rosenwaks, Z. Perspectives on the assessment of human sperm chromatin integrity. Fertil. Steril. 2014, 102, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.-T.; Wang, R.-X.; He, B.; Mo, W.-Y.; Huang, L.; Wang, S.-K.; Mao, X.-B.; Cheng, J.-P.; Huang, Y.-Y.; Liu, R. Effect of sperm DNA fragmentation on clinical outcomes for Chinese couples undergoing in vitro fertilization or intracytoplasmic sperm injection. J. Int. Med. Res. 2016, 44, 1283–1291. [Google Scholar] [CrossRef]
- Alvarez Sedó, C.; Bilinski, M.; Lorenzi, D.; Uriondo, H.; Noblía, F.; Longobucco, V.; Lagar, E.V.; Nodar, F. Effect of sperm DNA fragmentation on embryo development: Clinical and biological aspects. JBRA Assist. Reprod. 2017, 21, 343–350. [Google Scholar]
- Simon, L.; Emery, B.R.; Carrell, D.T. Review: Diagnosis and impact of sperm DNA alterations in assisted reproduction. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 44, 38–56. [Google Scholar] [CrossRef]
- Colaco, S.; Sakkas, D. Paternal factors contributing to embryo quality. J. Assist. Reprod. Genet. 2018, 35, 1953–1968. [Google Scholar] [CrossRef]
- Carrell, D.T.; Emery, B.R.; Hammoud, S. Altered protamine expression and diminished spermatogenesis: What is the link? Hum. Reprod. Update 2007, 13, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Imken, L.; Rouba, H.; El Houate, B.; Louanjli, N.; Barakat, A.; Chafik, A.; McElreavey, K. Mutations in the protamine locus: Association with spermatogenic failure? Mol. Hum. Reprod. 2009, 15, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, D.; Brauner, R.; Lin, L.; De Perdigo, A.; Weryha, G.; Muresan, M.; Boudjenah, R.; Guerra-Junior, G.; Maciel-Guerra, A.T.; Achermann, J.C.; et al. Mutations in NR5A1 associated with ovarian insufficiency. N. Engl. J. Med. 2009, 360, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Bashamboo, A.; Ferraz-de-Souza, B.; Lourenço, D.; Lin, L.; Sebire, N.J.; Montjean, D.; Bignon-Topalovic, J.; Mandelbaum, J.; Siffroi, J.P.; Christin-Maitre, S.; et al. Human Male Infertility Associated with Mutations in NR5A1 Encoding Steroidogenic Factor 1. Am. J. Hum. Genet. 2010, 87, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Mason, J.B. Folate and colorectal carcinogenesis: Is DNA repair the missing link? Am. J. Gastroenterol. 1998, 93, 2013–2016. [Google Scholar] [CrossRef]
- Ebisch, I.M.W.; van Heerde, W.L.; Thomas, C.M.G.; van der Put, N.; Wong, W.Y.; Steegers-Theunissen, R.P.M. C677T methylenetetrahydrofolate reductase polymorphism interferes with the effects of folic acid and zinc sulfate on sperm concentration. Fertil. Steril. 2003, 80, 1190–1194. [Google Scholar] [CrossRef]
- Montjean, D.; Benkhalifa, M.; Dessolle, L.; Cohen-Bacrie, P.; Belloc, S.; Siffroi, J.-P.; Ravel, C.; Bashamboo, A.; McElreavey, K. Polymorphisms in MTHFR and MTRR genes associated with blood plasma homocysteine concentration and sperm counts. Fertil. Steril. 2011, 95, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-G.; Ding, X.-F.; Liao, A.-H.; Kong, X.-B.; Xiong, C.-L. Expression of CatSper family transcripts in the mouse testis during post-natal development and human ejaculated spermatozoa: Relationship to sperm motility. Mol. Hum. Reprod. 2007, 13, 299–306. [Google Scholar] [CrossRef]
- Zuccarello, D.; Ferlin, A.; Cazzadore, C.; Pepe, A.; Garolla, A.; Moretti, A.; Cordeschi, G.; Francavilla, S.; Foresta, C. Mutations in dynein genes in patients affected by isolated non-syndromic asthenozoospermia. Hum. Reprod. 2008, 23, 1957–1962. [Google Scholar] [CrossRef]
- Dieterich, K.; Soto Rifo, R.; Faure, A.K.; Hennebicq, S.; Ben Amar, B.; Zahi, M.; Perrin, J.; Martinez, D.; Sèle, B.; Jouk, P.-S.; et al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat. Genet. 2007, 39, 661–665. [Google Scholar] [CrossRef]
- Dam, A.H.D.M.; Koscinski, I.; Kremer, J.A.M.; Moutou, C.; Jaeger, A.-S.; Oudakker, A.R.; Tournaye, H.; Charlet, N.; Lagier-Tourenne, C.; van Bokhoven, H.; et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am. J. Hum. Genet. 2007, 81, 813–820. [Google Scholar] [CrossRef]
- Harbuz, R.; Zouari, R.; Pierre, V.; Ben Khelifa, M.; Kharouf, M.; Coutton, C.; Merdassi, G.; Abada, F.; Escoffier, J.; Nikas, Y.; et al. A Recurrent Deletion of DPY19L2 Causes Infertility in Man by Blocking Sperm Head Elongation and Acrosome Formation. Am. J. Hum. Genet. 2011, 88, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Grasa, P.; Coward, K.; Young, C.; Parrington, J. The pattern of localization of the putative oocyte activation factor, phospholipase Czeta, in uncapacitated, capacitated, and ionophore-treated human spermatozoa. Hum. Reprod. 2008, 23, 2513–2522. [Google Scholar] [CrossRef] [PubMed]
- Nomikos, M.; Kashir, J.; Swann, K.; Lai, F.A. Sperm PLCζ: From structure to Ca2+ oscillations, egg activation and therapeutic potential. FEBS Lett. 2013, 587, 3609–3616. [Google Scholar] [CrossRef]
- Saleh, A.; Kashir, J.; Thanassoulas, A.; Safieh-Garabedian, B.; Lai, F.A.; Nomikos, M. Essential Role of Sperm-Specific PLC-Zeta in Egg Activation and Male Factor Infertility: An Update. Front. Cell Dev. Biol. 2020, 8, 28. [Google Scholar] [CrossRef]
- Nozawa, K.; Satouh, Y.; Fujimoto, T.; Oji, A.; Ikawa, M. Sperm-borne phospholipase C zeta-1 ensures monospermic fertilization in mice. Sci. Rep. 2018, 8, 1315. [Google Scholar] [CrossRef]
- Saunders, C.M.; Larman, M.G.; Parrington, J.; Cox, L.J.; Royse, J.; Blayney, L.M.; Swann, K.; Lai, F.A. PLC zeta: A sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development 2002, 129, 3533–3544. [Google Scholar] [CrossRef] [PubMed]
- Kouchi, Z.; Fukami, K.; Shikano, T.; Oda, S.; Nakamura, Y.; Takenawa, T.; Miyazaki, S. Recombinant phospholipase Czeta has high Ca2+ sensitivity and induces Ca2+ oscillations in mouse eggs. J. Biol. Chem. 2004, 279, 10408–10412. [Google Scholar] [CrossRef]
- Yoon, S.-Y.; Jellerette, T.; Salicioni, A.M.; Lee, H.C.; Yoo, M.-S.; Coward, K.; Parrington, J.; Grow, D.; Cibelli, J.B.; Visconti, P.E.; et al. Human sperm devoid of PLC, zeta 1 fail to induce Ca(2+) release and are unable to initiate the first step of embryo development. J. Clin. Invest. 2008, 118, 3671–3681. [Google Scholar] [CrossRef]
- Ebner, T.; Montag, M.; Van der Ven, K.; Van der Ven, H.; Shebl, O.; Oppelt, P.; Hirchenhain, J.; Krüssel, J.; Maxrath, B.; Gnoth, C.; et al. Live birth after artificial oocyte activation using a ready-to-use ionophore: A prospective multicentre study. Reprod. Biomed. Online 2015, 30, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Bonte, D.; Ferrer-Buitrago, M.; Dhaenens, L.; Popovic, M.; Thys, V.; De Croo, I.; De Gheselle, S.; Steyaert, N.; Boel, A.; Meerschaut, F.V.; et al. Assisted oocyte activation significantly increases fertilization and pregnancy outcome in patients with low and total failed fertilization after intracytoplasmic sperm injection: A 17-year retrospective study. Fertil. Steril. 2019, 112, 266–274. [Google Scholar] [CrossRef]
- Nazarian, H.; Azad, N.; Nazari, L.; Piryaei, A.; Heidari, M.H.; Masteri-Farahani, R.; Karimi, M.; Ghaffari-Novin, M. Effect of Artificial Oocyte Activation on Intra-Cytoplasmic Sperm Injection Outcomes in Patients with Lower Percentage of Sperm Containing Phospholipase Cζ: A Randomized Clinical Trial. J. Reprod. Infertil. 2019, 20, 3–9. [Google Scholar] [PubMed]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 2007, 19, 266–272. [Google Scholar] [CrossRef]
- Miller, D.; Brinkworth, M.; Iles, D. Paternal DNA packaging in spermatozoa: More than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 2010, 139, 287–301. [Google Scholar] [CrossRef]
- Steilmann, C.; Paradowska, A.; Bartkuhn, M.; Vieweg, M.; Schuppe, H.C.; Bergmann, M.; Kliesch, S.; Weidner, W.; Steger, K. Presence of histone H3 acetylated at lysine 9 in male germ cells and its distribution pattern in the genome of human spermatozoa. Reprod. Fertil. Dev. 2011, 23, 997–1011. [Google Scholar] [CrossRef]
- Sonnack, V.; Failing, K.; Bergmann, M.; Steger, K. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia 2002, 34, 384–390. [Google Scholar] [CrossRef]
- Schon, S.B.; Luense, L.J.; Wang, X.; Bartolomei, M.S.; Coutifaris, C.; Garcia, B.A.; Berger, S.L. Histone modification signatures in human sperm distinguish clinical abnormalities. Assist. Reprod. Genet. 2019, 36, 267–275. [Google Scholar] [CrossRef]
- Štiavnická, M.; García-Álvarez, O.; Ulčová-Gallová, Z.; Sutovsky, P.; Abril-Parreño, L.; Dolejšová, M.; Řimnáčová, H.; Moravec, J.; Hošek, P.; Lošan, P.; et al. H3K4me2 accompanies chromatin immaturity in human spermatozoa: An epigenetic marker for sperm quality assessment. Syst. Biol. Reprod. Med. 2019, 66, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Siklenka, K.; Erkek, S.; Godmann, M.; Lambrot, R.; McGraw, S.; Lafleur, C.; Cohen, T.; Xia, J.; Suderman, M.; Hallett, M.; et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015, 350, aab2006. [Google Scholar] [CrossRef] [PubMed]
- Moritz, L.; Hammoud, S.S. The Art of Packaging the Sperm Genome: Molecular and Structural Basis of the Histone-To-Protamine Exchange. Front. Endocrinol. 2022, 13, 895502. [Google Scholar] [CrossRef]
- Ravel, C.; Chantot-Bastaraud, S.; El Houate, B.; Berthaut, I.; Verstraete, L.; De Larouziere, V.; Lourenço, D.; Dumaine, A.; Antoine, J.M.; Mandelbaum, J.; et al. Mutations in the protamine 1 gene associated with male infertility. Mol. Hum. Reprod. 2007, 461–464. [Google Scholar] [CrossRef]
- Bai, H.; Sha, Y.; Tan, Y.; Li, P.; Zhang, Y.; Xu, J.; Xu, S.; Ji, Z.; Wang, X.; Chen, W.; et al. Deleterious variants in TAF7L cause human oligoasthenoteratozoospermia and its impairing histone to protamine exchange inducing reduced in vitro fertilization. Front. Endocrinol. (Lausanne) 2023, 13, 1099270. [Google Scholar] [CrossRef]
- Ostermeier, G.C.; Goodrich, R.J.; Moldenhauer, J.S.; Diamond, M.P.; Krawetz, S.A. A suite of novel human spermatozoal RNAs. J. Androl. 2005, 26, 70–74. [Google Scholar] [CrossRef]
- He, L.; Vasiliou, K.; Nebert, D.W. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum. Genom. 2009, 3, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Chioccarelli, T.; Manfrevola, F.; Ferraro, B.; Sellitto, C.; Cobellis, G.; Migliaccio, M.; Fasano, S.; Pierantoni, R.; Chianese, R. Expression Patterns of Circular RNAs in High Quality and Poor Quality Human Spermatozoa. Front. Endocrinol. 2019, 10, 435. [Google Scholar] [CrossRef]
- Shi, Z.; Yu, M.; Guo, T.; Sui, Y.; Tian, Z.; Ni, X.; Chen, X.; Jiang, M.; Jiang, J.; Lu, Y.; et al. MicroRNAs in spermatogenesis dysfunction and male infertility: Clinical phenotypes, mechanisms and potential diagnostic biomarkers. Front. Endocrinol. (Lausanne) 2024, 15, 1293368. [Google Scholar] [CrossRef]
- Doerksen, T.; Trasler, J.M. Developmental exposure of male germ cells to 5-azacytidine results in abnormal preimplantation development in rats. Biol. Reprod. 1996, 55, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.J.; Carvalho, F.; Sousa, M.; Barros, A. Genomic imprinting in disruptive spermatogenesis. Lancet 2004, 363, 1700–1702. [Google Scholar] [CrossRef] [PubMed]
- Houshdaran, S.; Cortessis, V.K.; Siegmund, K.; Yang, A.; Laird, P.W.; Sokol, R.Z. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS ONE 2007, 2, e1289. [Google Scholar] [CrossRef]
- Kobayashi, H.; Sato, A.; Otsu, E.; Hiura, H.; Tomatsu, C.; Utsunomiya, T.; Sasaki, H.; Yaegashi, N.; Arima, T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum. Mol. Genet. 2007, 16, 2542–2551. [Google Scholar] [CrossRef]
- Marques, C.J.; Costa, P.; Vaz, B.; Carvalho, F.; Fernandes, S.; Barros, A.; Sousa, M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol. Hum. Reprod. 2008, 14, 67–74. [Google Scholar] [CrossRef]
- Boissonnas, C.C.; Abdalaoui, H.E.; Haelewyn, V.; Fauque, P.; Dupont, J.M.; Gut, I.; Vaiman, D.; Jouannet, P.; Tost, J.; Jammes, H. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur. J. Hum. Genet. 2010, 18, 73–80. [Google Scholar] [CrossRef]
- Montjean, D.; Ravel, C.; Benkhalifa, M.; Cohen-Bacrie, P.; Berthaut, I.; Bashamboo, A.; McElreavey, K. Methylation changes in mature sperm deoxyribonucleic acid from oligozoospermic men: Assessment of genetic variants and assisted reproductive technology outcome. Fertil. Steril. 2013, 100, 1241–1247. [Google Scholar] [CrossRef]
- Kuhtz, J.; Schneider, E.; El Hajj, N.; Zimmermann, L.; Fust, O.; Linek, B.; Seufert, R.; Hahn, T.; Schorsch, M.; Haaf, T. Epigenetic heterogeneity of developmentally important genes in human sperm: Implications for assisted reproduction outcome. Epigenetics 2015, 9, 1648–1658. [Google Scholar] [CrossRef]
- Laurentino, S.; Beygo, J.; Nordhoff, V.; Kliesch, S.; Wistuba, J.; Borgmann, J.; Buiting, K.; Horsthemke, B.; Gromoll, J. Epigenetic germline mosaicism in infertile men. Hum. Mol. Genet. 2015, 24, 1295–1304. [Google Scholar] [CrossRef]
- Montjean, D.; Zini, A.; Ravel, C.; Belloc, S.; Dalleac, A.; Copin, H.; Boyer, P.; McElreavey, K.; Benkhalifa, M. Sperm global DNA methylation level: Association with semen parameters and genome integrity. Andrology 2015, 3, 235–240. [Google Scholar] [CrossRef]
- Urdinguio, R.G.; Bayón, G.F.; Dmitrijeva, M.; Toraño, E.G.; Bravo, C.; Fraga, M.F.; Bassas, L.; Larriba, S.; Fernández, A.F. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum. Reprod. 2015, 30, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Anway, M.D.; Cupp, A.S.; Uzumcu, M.; Skinner, M.K. Epigenetic Transgenerational Actions of Endocrine Disruptors and Male Fertility. Science 2005, 308, 1466–1469. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Skinner, M.K. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE 2013, 8, e55387. [Google Scholar]
- Liehr, T. Chapter 14—Nuclear architecture. In Cytogenomics [Internet]; Liehr, T., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 297–305. Available online: https://www.sciencedirect.com/science/article/pii/B9780128235799000114 (accessed on 16 January 2024).
- Manvelyan, M.; Hunstig, F.; Bhatt, S.; Mrasek, K.; Pellestor, F.; Weise, A.; Simonyan, I.; Aroutiounian, R.; Liehr, T. Chromosome distribution in human sperm—A 3D multicolor banding-study. Mol. Cytogenet. 2008, 1, 25. [Google Scholar] [CrossRef]
- Karamysheva, T.; Kosyakova, N.; Guediche, N.; Liehr, T. Small supernumerary marker chromosomes and the nuclear architecture of sperm—A study in a fertile and an infertile brother. Syst. Biol. Reprod. Med. 2015, 61, 32–36. [Google Scholar] [CrossRef]
- Liehr, T.; Hamid Al-Rikabi, A.B. Impaired Spermatogenesis due to Small Supernumerary Marker Chromosomes: The Reason for Infertility Is Only Reliably Ascertainable by Cytogenetics. Sex. Dev. 2018, 12, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-G.; Huang, S.-Y.; Zhou, H.; Liao, A.-H.; Xiong, C.-L. Quick recovery and characterization of cell-free DNA in seminal plasma of normozoospermia and azoospermia: Implications for non-invasive genetic utilities. Asian J. Androl. 2009, 11, 703–709. [Google Scholar] [CrossRef]
- Di Pizio, P.; Celton, N.; Menoud, P.A.; Belloc, S.; Cohen Bacrie, M.; Belhadri-Mansouri, N.; Rives, N.; Cabry, R.; Benkhalifa, M. Seminal cell-free DNA and sperm characteristic’s: An added biomarker for male infertility investigation. Andrologia 2021, 53, e13822. [Google Scholar] [CrossRef]
- Vermeiden, J.P.W.; Bernardus, R.E. Are imprinting disorders more prevalent after human in vitro fertilization or intracytoplasmic sperm injection? Fertil. Steril. 2013, 99, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Q.; Wu, L.; Wang, H.; Shi, H.; Yang, C.; Gu, Y.; Li, J.; Ji, Z. SperMD: The expression atlas of sperm maturation. BMC Bioinform. 2024, 25, 29. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montjean, D.; Beaumont, M.; Natiq, A.; Louanjli, N.; Hazout, A.; Miron, P.; Liehr, T.; Cabry, R.; Ratbi, I.; Benkhalifa, M. Genome and Epigenome Disorders and Male Infertility: Feedback from 15 Years of Clinical and Research Experience. Genes 2024, 15, 377. https://doi.org/10.3390/genes15030377
Montjean D, Beaumont M, Natiq A, Louanjli N, Hazout A, Miron P, Liehr T, Cabry R, Ratbi I, Benkhalifa M. Genome and Epigenome Disorders and Male Infertility: Feedback from 15 Years of Clinical and Research Experience. Genes. 2024; 15(3):377. https://doi.org/10.3390/genes15030377
Chicago/Turabian StyleMontjean, Debbie, Marion Beaumont, Abdelhafid Natiq, Noureddine Louanjli, Andre Hazout, Pierre Miron, Thomas Liehr, Rosalie Cabry, Ilham Ratbi, and Moncef Benkhalifa. 2024. "Genome and Epigenome Disorders and Male Infertility: Feedback from 15 Years of Clinical and Research Experience" Genes 15, no. 3: 377. https://doi.org/10.3390/genes15030377
APA StyleMontjean, D., Beaumont, M., Natiq, A., Louanjli, N., Hazout, A., Miron, P., Liehr, T., Cabry, R., Ratbi, I., & Benkhalifa, M. (2024). Genome and Epigenome Disorders and Male Infertility: Feedback from 15 Years of Clinical and Research Experience. Genes, 15(3), 377. https://doi.org/10.3390/genes15030377






