Enhancing Variant Prioritization in VarFish through On-Premise Computational Facial Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview
2.2. Cohort Description
2.3. Prioritization Approaches
2.4. Step-by-Step Setup
- Initialization of Services: VarFish, CADD, CADA, GestaltMatcher, and PEDIA are initiated as separate web services. For instance, these services can be initialized on the same machine but on different ports. Instructions for starting each service can be found in the Supplementary Materials.
- Configuration in VarFish: VarFish’s settings file is configured to include the URLs for the CADD, CADA, GestaltMatcher, and PEDIA web services. This ensures seamless communication and interaction between VarFish and the aforementioned tools. These tools can be hosted either in the same machine for the on-premise solution or accessible via the web services provided by the inventors.
- Integration of GestaltMatcher into Prioritization:
- 3.1.
- The user activates GestaltMatcher within VarFish. The face sender module from the PEDIA middleware is embedded as an iFrame in the prioritization page of VarFish.
- 3.2.
- Upon selecting the frontal image of the patient for the case and submitting it, the image is transmitted via the POST method exposed by the REST API endpoint of the GestaltMatcher web service.
- 3.3.
- After receiving a successful response from GestaltMatcher, the suggested gene list along with scores is relayed to the parent window of VarFish.
- 3.4.
- Additionally, the file name of the last photo successfully submitted to GestaltMatcher is transmitted back to VarFish. This communication from the embedded child frame to the parent window is facilitated using the window.postMessage method.
- 3.5.
- A listener is incorporated into the prioritization page of VarFish to capture message events sent from the iFrame. The received data are then stored in the variant query store of VarFish. This process ensures that the patient image does not require re-uploading when the case is reopened in VarFish, as it only needs to be submitted once per case.
- 3.6.
- Subsequently, when the user performs filtering, the resulting variants table displays the Gestalt scores obtained from the last image submitted to GestaltMatcher.
- Enabling PEDIA-based prioritization: within the prioritization page of VarFish, it automatically triggers phenotype-based prioritization using the CADA algorithm. Furthermore, it activates variant pathogenicity-based prioritization utilizing CADD scores, which predict the deleteriousness of the variants.
2.5. Automation
3. Results
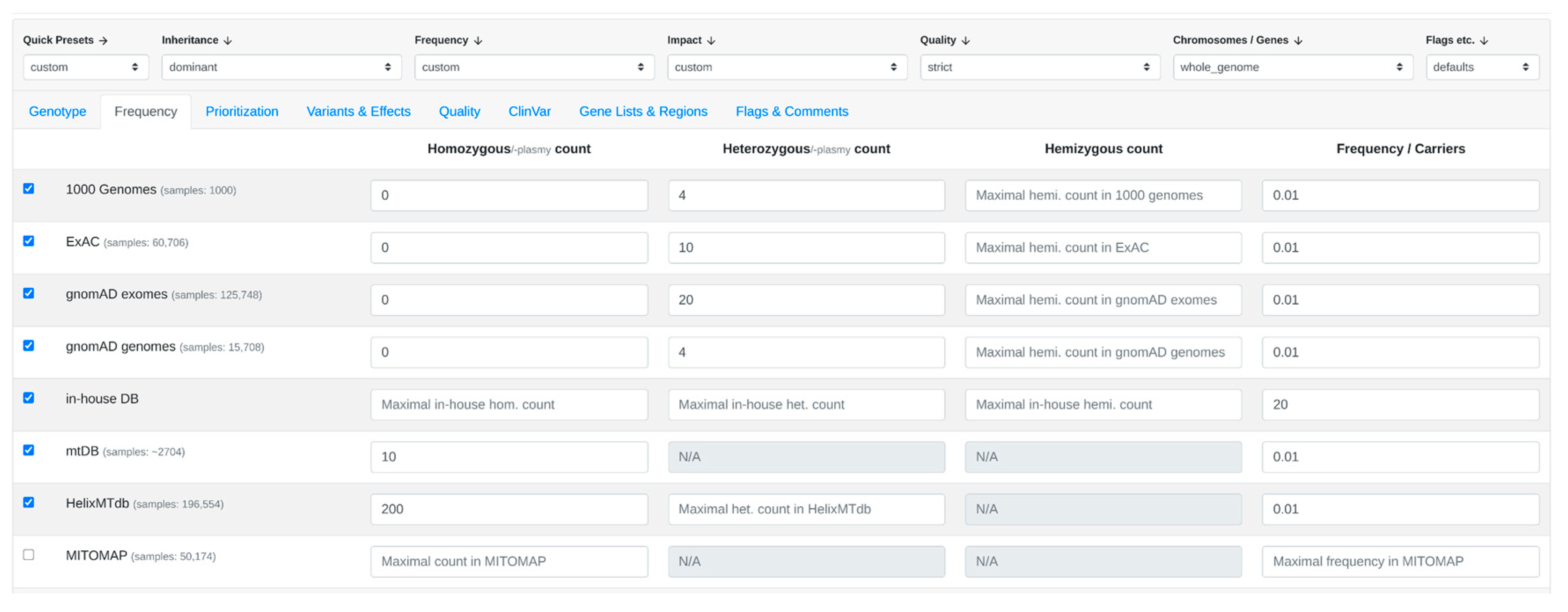
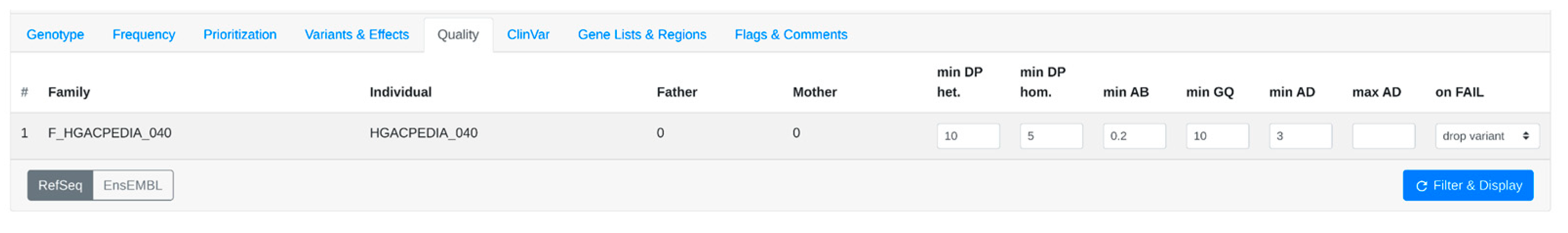
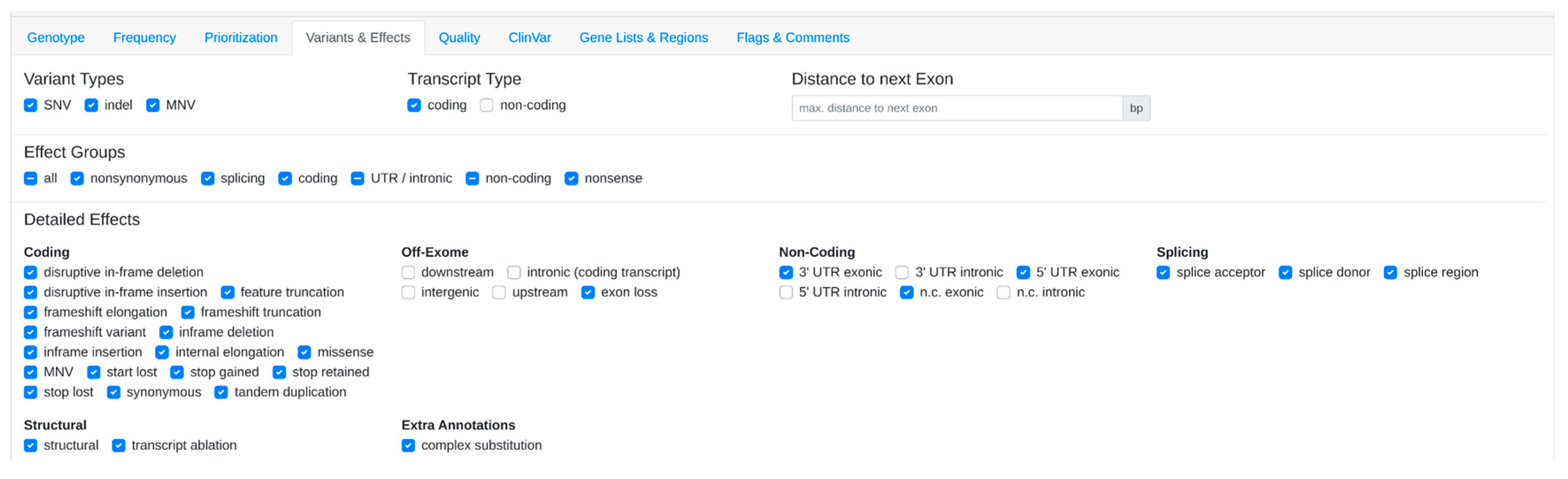
3.1. Step-by-Step Analysis
- 2.
- CADA scores are acquired from the CADA web service by transmitting clinical features in HPO terms.
- 3.
- Subsequently, CADD scores are obtained from the CADD web service by forwarding the filtered variants. The highest CADD score is chosen for each gene.
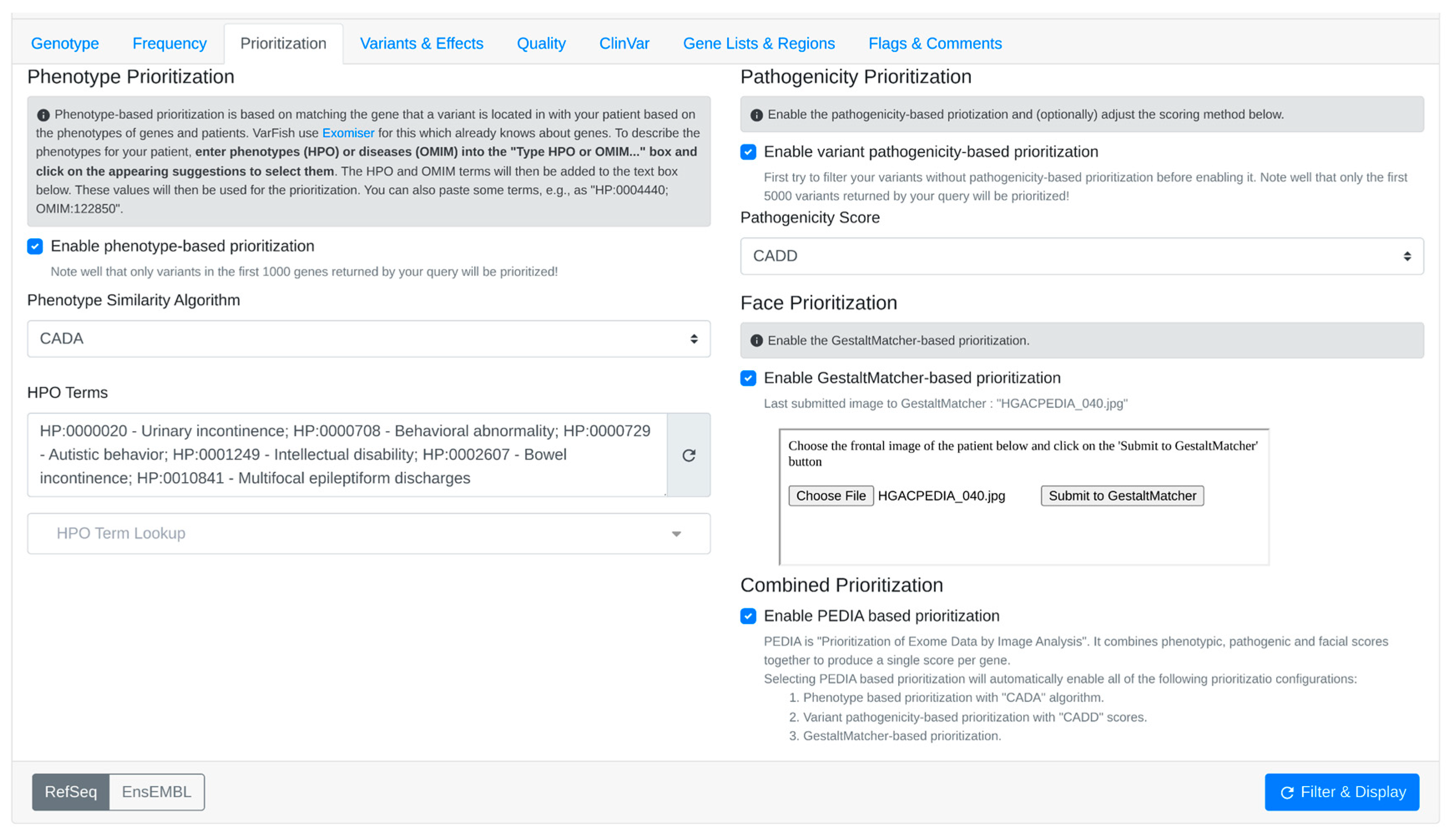
- 4.
- Upload the patient’s facial image to obtain Gestalt scores calculated by GestaltMatcher (Figure 5).
- 5.
- After checking the “Enable PEDIA-based prioritization” button and clicking “Filter & Display,” these scores (CADA, CADD, and Gestalt) are then dispatched to the PEDIA web service via the REST API endpoint to procure PEDIA scores per gene.
- 6.
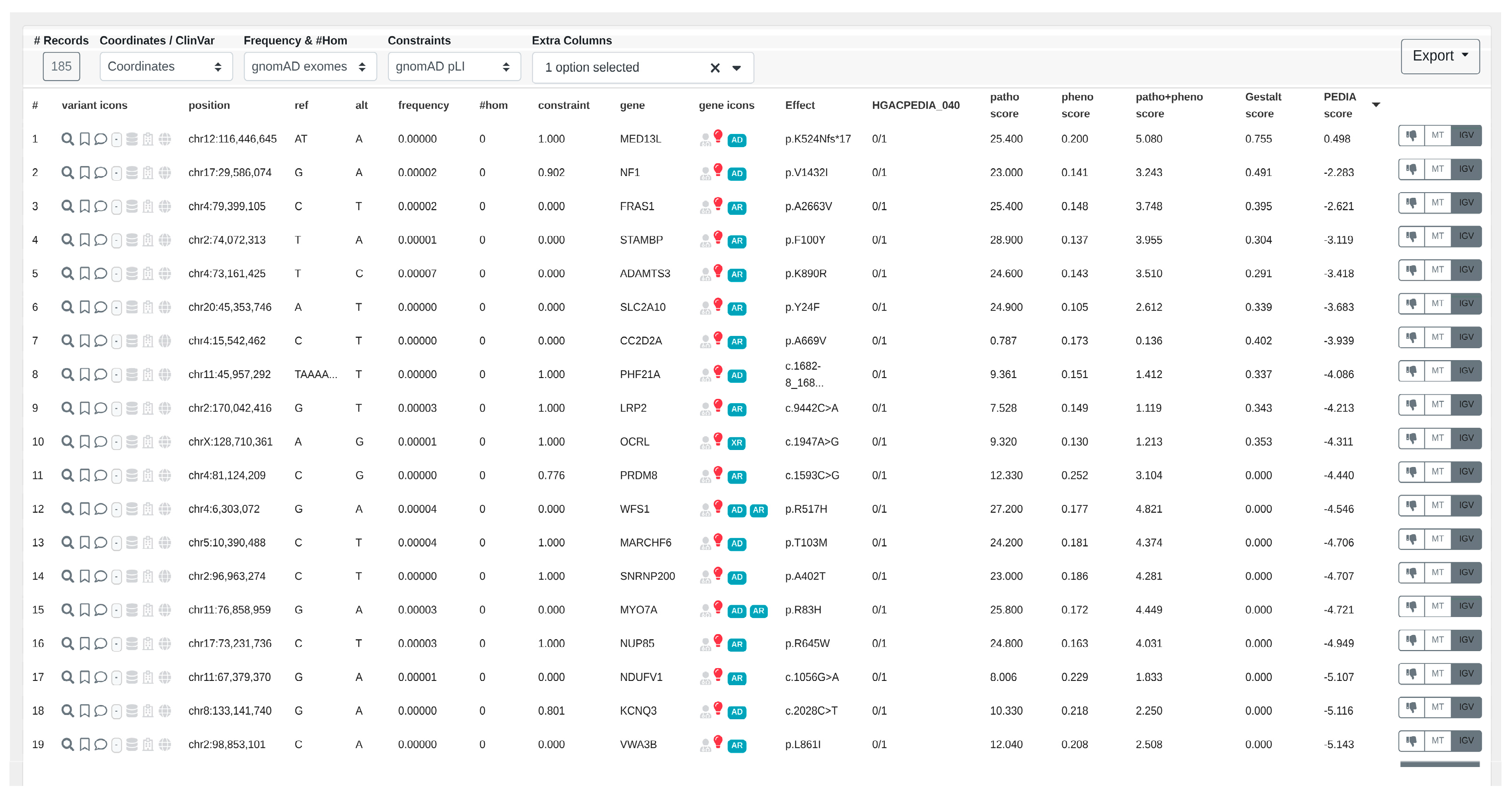
- In the resulting variants table (Figure 6), PEDIA scores are displayed in a distinct column alongside CADA, CADD, and Gestalt scores. Variants associated with genes having higher PEDIA scores are prioritized accordingly.
3.2. Visualizing Results in VarFish
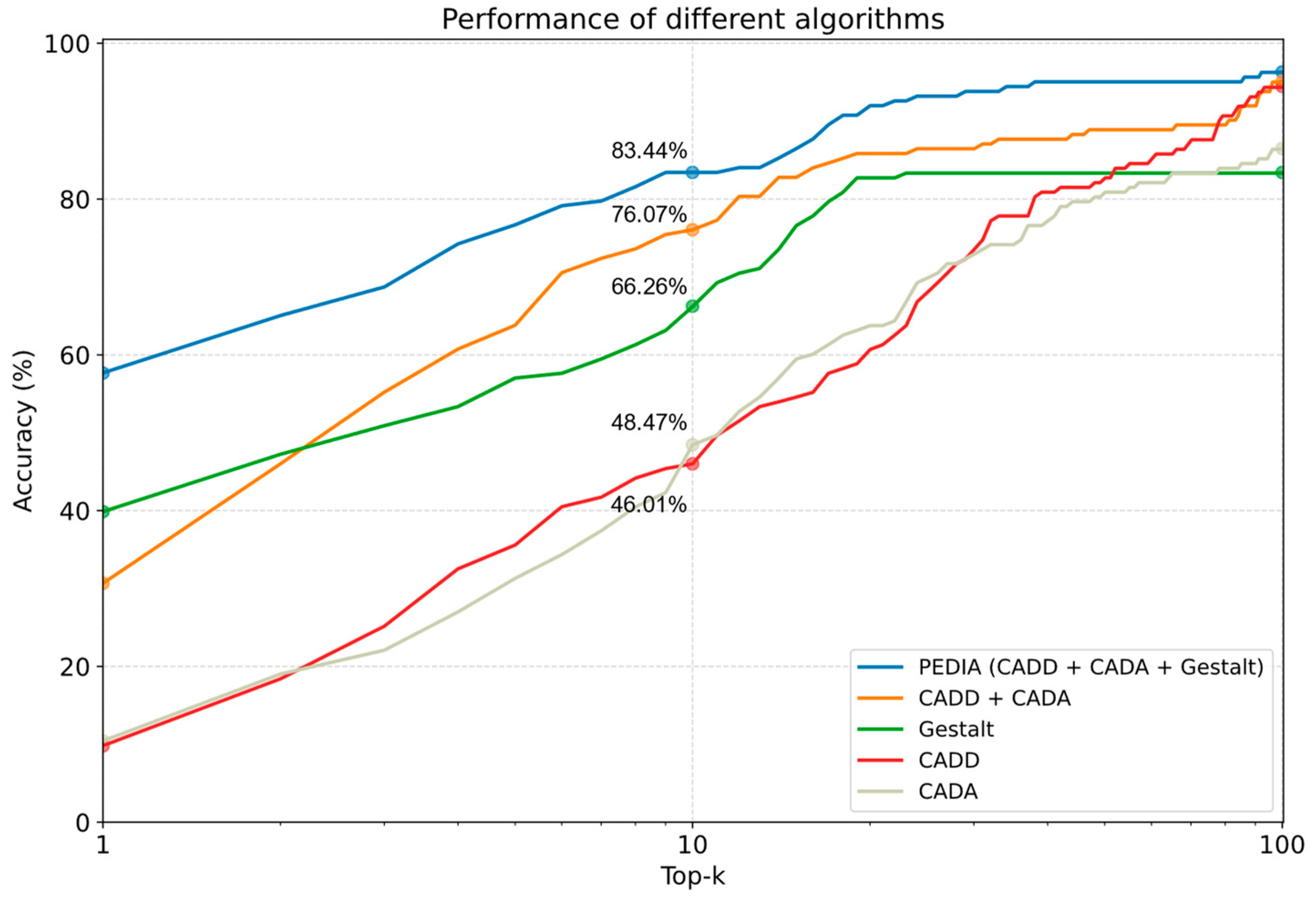
3.3. Performance Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferreira, C.R. The burden of rare diseases. Am. J. Med. Genet. A 2019, 179, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.C.; Leung, G.K.; Mak, C.C.; Fung, J.L.; Lee, M.; Pei, S.L.; Yu, M.H.; Hui, V.C.; Chan, J.C.; Chau, J.F.; et al. Rapid whole-exome sequencing facilitates precision medicine in paediatric rare disease patients and reduces healthcare costs. Lancet Reg. Health West. Pac. 2020, 1, 100001. [Google Scholar] [CrossRef] [PubMed]
- Köhler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, S.; Baynam, G.; Brower, A.M.; et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021, 49, D1207–D1217. [Google Scholar] [CrossRef] [PubMed]
- Köhler, S.; Schulz, M.H.; Krawitz, P.; Bauer, S.; Dölken, S.; Ott, C.E.; Mundlos, C.; Horn, D.; Mundlos, S.; Robinson, P.N. Clinical Diagnostics in Human Genetics with Semantic Similarity Searches in Ontologies. Am. J. Hum. Genet. 2009, 85, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Köhler, S.; Schulz, M.H.; Robinson, P.N. Bayesian ontology querying for accurate and noise-tolerant semantic searches. Bioinformatics 2012, 28, 2502–2508. [Google Scholar] [CrossRef] [PubMed]
- Smedley, D.; Jacobsen, J.O.B.; Jäger, M.; Köhler, S.; Holtgrewe, M.; Schubach, M.; Siragusa, E.; Zemojtel, T.; Buske, O.J.; Washington, N.L.; et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc. 2015, 10, 2004–2015. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Robinson, P.N.; Wang, K. Phenolyzer: Phenotype-based prioritization of candidate genes for human diseases. Nat. Methods 2015, 12, 841–843. [Google Scholar] [CrossRef]
- Jagadeesh, K.A.; Birgmeier, J.; Guturu, H.; Deisseroth, C.A.; Wenger, A.M.; Bernstein, J.A.; Bejerano, G. Phrank measures phenotype sets similarity to greatly improve Mendelian diagnostic disease prioritization. Genet. Med. 2019, 21, 464–470. [Google Scholar] [CrossRef]
- Birgmeier, J.; Haeussler, M.; Deisseroth, C.A.; Steinberg, E.H.; Jagadeesh, K.A.; Ratner, A.J.; Guturu, H.; Wenger, A.M.; Diekhans, M.E.; Stenson, P.D.; et al. AMELIE speeds Mendelian diagnosis by matching patient phenotype and genotype to primary literature. Sci. Transl. Med. 2020, 12, eaau9113. [Google Scholar] [CrossRef]
- Zhao, M.; Havrilla, J.M.; Fang, L.; Chen, Y.; Peng, J.; Liu, C.; Wu, C.; Sarmady, M.; Botas, P.; Isla, J.; et al. Phen2Gene: Rapid phenotype-driven gene prioritization for rare diseases. NAR Genom. Bioinform. 2020, 2, lqaa032. [Google Scholar] [CrossRef]
- Robinson, P.N.; Ravanmehr, V.; Jacobsen, J.O.; Danis, D.; Zhang, X.A.; Carmody, L.C.; Gargano, M.A.; Thaxton, C.L.; Karlebach, G.; Reese, J.; et al. Interpretable Clinical Genomics with a Likelihood Ratio Paradigm. Am. J. Hum. Genet. 2020, 107, 403–417. [Google Scholar] [CrossRef]
- Peng, C.; Dieck, S.; Schmid, A.; Ahmad, A.; Knaus, A.; Wenzel, M.; Mehnert, L.; Zirn, B.; Haack, T.; Ossowski, S.; et al. CADA: Phenotype-driven gene prioritization based on a case-enriched knowledge graph. NAR Genom. Bioinform. 2021, 3, lqab078. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, Y.; Yang, Y.; Huang, Y.; Zhao, S.; Zhao, H.; Yu, C.; Dong, X.; Zhang, Y.; Wang, L.; et al. PhenoApt leverages clinical expertise to prioritize candidate genes via machine learning. Am. J. Hum. Genet. 2022, 109, 270–281. [Google Scholar] [CrossRef]
- Kelly, C.; Szabo, A.; Pontikos, N.; Arno, G.; Robinson, P.N.; Jacobsen, J.O.; Smedley, D.; Cipriani, V. Phenotype-aware prioritisation of rare Mendelian disease variants. Trends Genet. 2022, 38, 1271–1283. [Google Scholar] [CrossRef]
- Zhai, W.; Huang, X.; Shen, N.; Zhu, S. Phen2Disease: A phenotype-driven model for disease and gene prioritization by bidirectional maximum matching semantic similarities. Brief. Bioinform. 2023, 24, bbad172. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, C.; Deng, W.; Wu, D.; Weng, C.; Zhou, Y.; Wang, K. Enhancing phenotype recognition in clinical notes using large language models: PhenoBCBERT and PhenoGPT. Patterns 2024, 5, 100887. [Google Scholar] [CrossRef] [PubMed]
- Dudding-Byth, T.; Baxter, A.; Holliday, E.G.; Hackett, A.; O’donnell, S.; White, S.M.; Attia, J.; Brunner, H.; De Vries, B.; Koolen, D.; et al. Computer face-matching technology using two-dimensional photographs accurately matches the facial gestalt of unrelated individuals with the same syndromic form of intellectual disability. BMC Biotechnol. 2017, 17, 90. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.; Gupta, T.; Saini, A.; Singh, P.; Balasubramanian, R. A Deep Learning Frame-Work for Recognizing Developmental Disorders. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; pp. 705–714. [Google Scholar]
- Liehr, T.; Acquarola, N.; Pyle, K.; St-Pierre, S.; Rinholm, M.; Bar, O.; Wilhelm, K.; Schreyer, I. Next generation phenotyping in Emanuel and Pallister-Killian syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin. Genet. 2018, 93, 378–381. [Google Scholar] [CrossRef] [PubMed]
- van der Donk, R.; Jansen, S.; Schuurs-Hoeijmakers, J.H.M.; Koolen, D.A.; Goltstein, L.C.M.J.; Hoischen, A.; Brunner, H.G.; Kemmeren, P.; Nellåker, C.; Vissers, L.E.L.M.; et al. Next-generation phenotyping using computer vision algorithms in rare genomic neurodevelopmental disorders. Genet. Med. 2019, 21, 1719–1725. [Google Scholar] [CrossRef]
- Gurovich, Y.; Hanani, Y.; Bar, O.; Nadav, G.; Fleischer, N.; Gelbman, D.; Basel-Salmon, L.; Krawitz, P.M.; Kamphausen, S.B.; Zenker, M.; et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019, 25, 60–64. [Google Scholar] [CrossRef]
- Liu, H.; Mo, Z.-H.; Yang, H.; Zhang, Z.-F.; Hong, D.; Wen, L.; Lin, M.-Y.; Zheng, Y.-Y.; Zhang, Z.-W.; Xu, X.-W.; et al. Automatic Facial Recognition of Williams-Beuren Syndrome Based on Deep Convolutional Neural Networks. Front. Pediatr. 2021, 9, 648255. [Google Scholar] [CrossRef]
- Porras, A.R.; Rosenbaum, K.; Tor-Diez, C.; Summar, M.; Linguraru, M.G. Development and evaluation of a machine learning-based point-of-care screening tool for genetic syndromes in children: A multinational retrospective study. Lancet Digit. Health 2021, 3, e635–e643. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Zheng, Y.-Y.; Xin, Y.; Sun, L.; Yang, H.; Lin, M.-Y.; Liu, C.; Li, B.-N.; Zhang, Z.-W.; Zhuang, J.; et al. Genetic syndromes screening by facial recognition technology: VGG-16 screening model construction and evaluation. Orphanet J. Rare Dis. 2021, 16, 344. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-C.; Bar-Haim, A.; Moosa, S.; Ehmke, N.; Gripp, K.W.; Pantel, J.T.; Danyel, M.; Mensah, M.A.; Horn, D.; Rosnev, S.; et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat. Genet. 2022, 54, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Danyel, M.; Grundmann, K.; Brunet, T.; Klinkhammer, H.; Hsieh, T.-C.; Engels, H.; Peters, S.; Knaus, A.; Moosa, S.; et al. Next-generation phenotyping integrated in a national framework for patients with ultra-rare disorders improves genetic diagnostics and yields new molecular findings. medRxiv 2023. [Google Scholar] [CrossRef]
- Hsieh, T.-C.; Mensah, M.A.; Pantel, J.T.; Aguilar, D.; Bar, O.; Bayat, A.; Becerra-Solano, L.; Bentzen, H.B.; Biskup, S.; Borisov, O.; et al. PEDIA: Prioritization of exome data by image analysis. Genet. Med. 2019, 21, 2807–2814. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-C.; Lesmann, H.; Krawitz, P.M. Facilitating the Molecular Diagnosis of Rare Genetic Disorders Through Facial Phenotypic Scores. Curr. Protoc. 2023, 3, e906. [Google Scholar] [CrossRef] [PubMed]
- Lesmann, H.; Lyon, G.J.; Caro, P.; Abdelrazek, I.M.; Moosa, S.; Pantel, J.T.; Klinkhammer, H.; Hagen, M.T.; Kamphans, T.; Meiswinkel, W.; et al. GestaltMatcher Database—A FAIR database for medical imaging data of rare disorders. medRxiv 2023. [Google Scholar] [CrossRef]
- Holtgrewe, M.; Stolpe, O.; Nieminen, M.; Mundlos, S.; Knaus, A.; Kornak, U.; Seelow, D.; Segebrecht, L.; Spielmann, M.; Fischer-Zirnsak, B.; et al. VarFish: Comprehensive DNA variant analysis for diagnostics and research. Nucleic Acids Res. 2020, 48, W162–W169. [Google Scholar] [CrossRef]
- Elbracht, M. GestaltMatcher Database Case. 7274. Available online: https://db.gestaltmatcher.org/id/7274 (accessed on 2 February 2024).
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Tavtigian, S.V.; Greenblatt, M.S.; Harrison, S.M.; Nussbaum, R.L.; Prabhu, S.A.; Boucher, K.M.; Biesecker, L.G. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018, 20, 1054–1060. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhasin, M.A.; Knaus, A.; Incardona, P.; Schmid, A.; Holtgrewe, M.; Elbracht, M.; Krawitz, P.M.; Hsieh, T.-C. Enhancing Variant Prioritization in VarFish through On-Premise Computational Facial Analysis. Genes 2024, 15, 370. https://doi.org/10.3390/genes15030370
Bhasin MA, Knaus A, Incardona P, Schmid A, Holtgrewe M, Elbracht M, Krawitz PM, Hsieh T-C. Enhancing Variant Prioritization in VarFish through On-Premise Computational Facial Analysis. Genes. 2024; 15(3):370. https://doi.org/10.3390/genes15030370
Chicago/Turabian StyleBhasin, Meghna Ahuja, Alexej Knaus, Pietro Incardona, Alexander Schmid, Manuel Holtgrewe, Miriam Elbracht, Peter M. Krawitz, and Tzung-Chien Hsieh. 2024. "Enhancing Variant Prioritization in VarFish through On-Premise Computational Facial Analysis" Genes 15, no. 3: 370. https://doi.org/10.3390/genes15030370
APA StyleBhasin, M. A., Knaus, A., Incardona, P., Schmid, A., Holtgrewe, M., Elbracht, M., Krawitz, P. M., & Hsieh, T.-C. (2024). Enhancing Variant Prioritization in VarFish through On-Premise Computational Facial Analysis. Genes, 15(3), 370. https://doi.org/10.3390/genes15030370






