Improving Genomic Predictions in Multi-Breed Cattle Populations: A Comparative Analysis of BayesR and GBLUP Models
Abstract
1. Introduction
2. Material and Methods
2.1. Ethics Statement
2.2. Animals and Phenotype
2.3. Genotyping Data
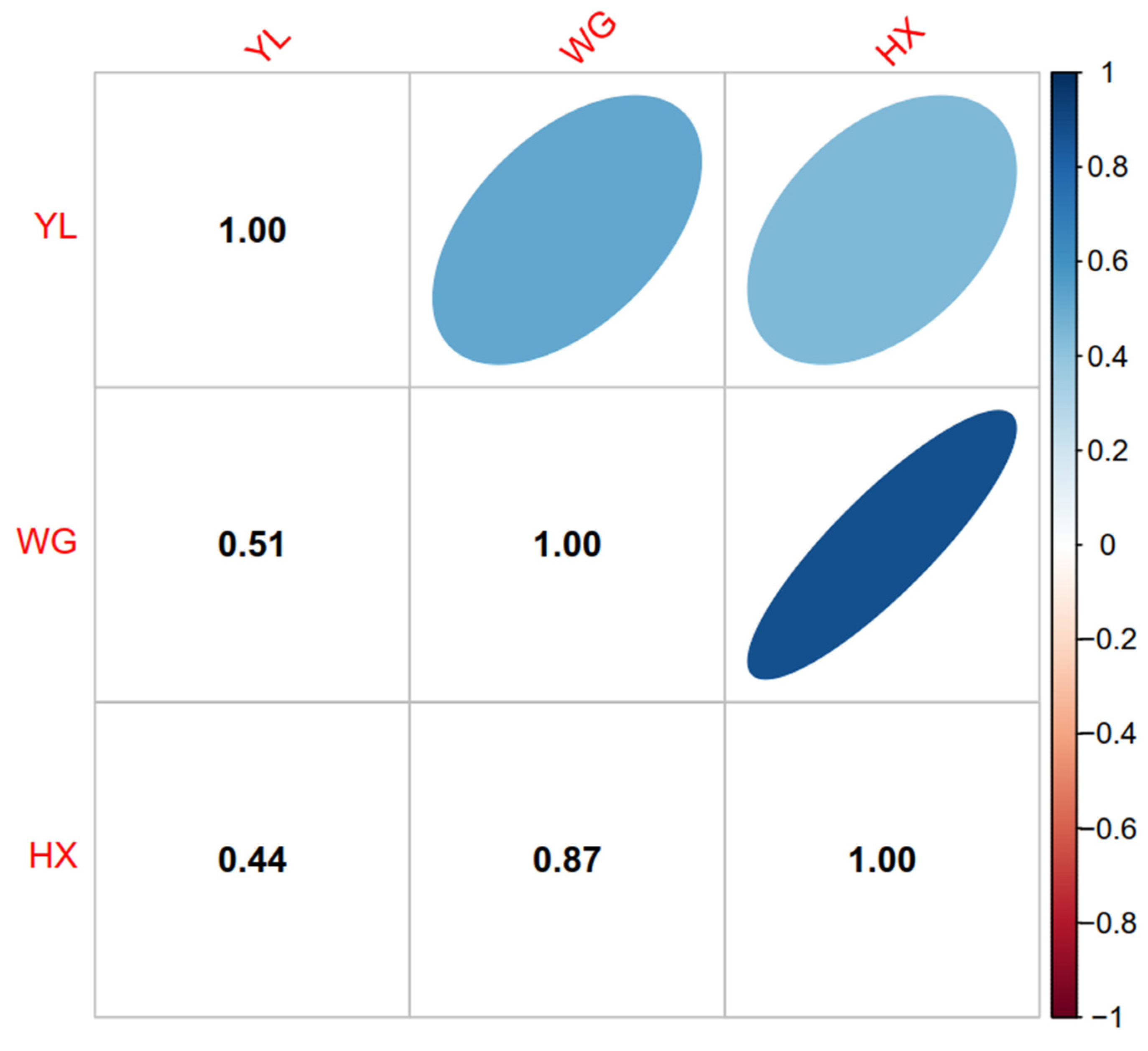
2.4. Genetic Correlations
2.5. Statistical Models
2.5.1. GBLUP Model
2.5.2. Multi-Breed GBLUP
2.5.3. Unweight G-Matrix ()
2.5.4. LD Weight G-Matrix ()
2.5.5. BayesR Model
3. Results
3.1. Descriptive Statistics of the Analyzed Traits
3.2. Genomic Relationship of Two Breeds
3.3. Prediction Accuracy of Within-Breed GS
3.4. Accuracy of Multi-Breed GS
4. Discussion
4.1. Impact of BayesR and GBLUP on GEBV in Single and Multiple Breed Genomic Selection
4.2. Effect of Using LD Information Weighted GRM on GEBV Accuracy
4.3. Influence of Genetic Correlation between Different Breeds on GEBV Accuracy
4.4. Impact of Marker Density on the Accuracy of Genomic Estimated Breeding Values
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNV | Copy number variant |
| CR | Call rate |
| EM | Expectation-maximization |
| GBLUP | Genome-enabled best linear unbiased prediction |
| GEBV | Genomic estimated breeding value |
| GP | Genomic prediction |
| GRM | Genomic relationship matrices |
| GS | Genomic selection |
| HD | High density |
| HX | Huaxi cattle |
| HW | Hardy–Weinberg Equilibrium |
| LD | Linkage disequilibrium |
| MAF | Minor allele frequencies |
| MCMC | Markov chain Monte Carlo |
| PCV | Phenotypic coefficient of variation |
| QTL | Quantitative trait locus |
| SNP | Single nucleotide polymorphism |
| SWT | Slaughter weight |
| WG | Chinese Wagyu cattle |
| WGS | Whole-genome sequencing |
| YL | Yunling cattle |
References
- Brøndum, R.F.; Su, G.; Lund, M.S.; Bowman, P.J.; Goddard, M.E.; Hayes, B.J. Genome position specific priors for genomic prediction. BMC Genom. 2012, 13, 543. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Bowman, P.J.; Chamberlain, A.C.; Verbyla, K.; Goddard, M.E. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 2009, 41, 51. [Google Scholar] [CrossRef] [PubMed]
- Brøndum, R.F.; Rius-Vilarrasa, E.; Strandén, I.; Su, G.; Guldbrandtsen, B.; Fikse, W.F.; Lund, M.S. Reliabilities of genomic prediction using combined reference data of the Nordic Red dairy cattle populations. J. Dairy Sci. 2011, 94, 4700–4707. [Google Scholar] [CrossRef] [PubMed]
- Iheshiulor, O.O.; Woolliams, J.A.; Yu, X.; Wellmann, R.; Meuwissen, T.H. Within- and across-breed genomic prediction using whole-genome sequence and single nucleotide polymorphism panels. Genet. Sel. Evol. 2016, 48, 15. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.S.; Roos, A.P.W.D.; Vries, A.G.D.; Druet, T.; Ducrocq, V.; Fritz, S.; Guillaume, F.; Guldbrandtsen, B.; Liu, Z.; Reents, R.; et al. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 2011, 43, 43. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.E.; Gredler, B.; Bolormaa, S.; Bowman, P.J.; Egger-Danner, C.; Fuerst, C.; Emmerling, R.; Sölkner, J.; Goddard, M.E.; Hayes, B.J. Short communication: Genomic selection using a multi-breed, across-country reference population. J. Dairy Sci. 2011, 94, 2625–2630. [Google Scholar] [CrossRef] [PubMed]
- Erbe, M.; Hayes, B.J.; Matukumalli, L.K.; Goswami, S.; Bowman, P.J.; Reich, C.M.; Mason, B.A.; Goddard, M.E. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 2012, 95, 4114–4129. [Google Scholar] [CrossRef]
- Kemper, K.E.; Reich, C.M.; Bowman, P.J.; Vander Jagt, C.J.; Chamberlain, A.J.; Mason, B.A.; Hayes, B.J.; Goddard, M.E. Improved precision of QTL mapping using a nonlinear Bayesian method in a multi-breed population leads to greater accuracy of across-breed genomic predictions. Genet. Sel. Evol. 2015, 47, 29. [Google Scholar] [CrossRef]
- Vanraden, P.M.; Null, D.J.; Sargolzaei, M.; Wiggans, G.R.; Tooker, M.E.; Cole, J.B.; Sonstegard, T.S.; Connor, E.E.; Winters, M.; van Kaam, J.B.C.H.; et al. Genomic imputation and evaluation using high-density Holstein genotypes. J. Dairy Sci. 2013, 96, 668–678. [Google Scholar] [CrossRef]
- Hozé, C.; Fritz, S.; Phocas, F.; Boichard, D.; Ducrocq, V.; Croiseau, P. Efficiency of multi-breed genomic selection for dairy cattle breeds with different sizes of reference population. J. Dairy Sci. 2014, 97, 3918–3929. [Google Scholar] [CrossRef]
- Meuwissen, T.; Goddard, M. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 2010, 185, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Calus, M.P.L.; Goddard, M.E.; Wientjes, Y.C.J.; Bowman, P.J.; Hayes, B.J. Multibreed genomic prediction using multitrait genomic residual maximum likelihood and multitask Bayesian variable selection. J. Dairy Sci. 2018, 101, 4279–4294. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.M.; Vanraden, P.M.; Tooker, M.E. Multibreed genomic evaluations using purebred Holsteins, Jerseys, and Brown Swiss. J. Dairy Sci. 2012, 95, 5378–5383. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, I.; Macleod, I.M.; Reich, C.M.; Breen, E.J.; Pryce, J.E. Optimizing genomic prediction for Australian Red dairy cattle. J. Dairy Sci. 2020, 103, 6276–6298. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, I.; Bowman, P.J.; Macleod, I.M.; Hayes, B.J.; Wang, T.; Bolormaa, S.; Goddard, M.E. Multi-breed genomic prediction using Bayes R with sequence data and dropping variants with a small effect. Genet. Sel. Evol. 2017, 49, 70. [Google Scholar] [CrossRef]
- Mohammad Rahimi, S.; Rashidi, A.; Esfandyari, H. Accounting for differences in linkage disequilibrium in multi-breed genomic prediction. Livest. Sci. 2020, 240, 104165. [Google Scholar] [CrossRef]
- Zhou, L.; Lund, M.S.; Wang, Y.; Su, G. Genomic predictions across Nordic Holstein and Nordic Red using the genomic best linear unbiased prediction model with different genomic relationship matrices. J. Anim. Breed. Genet. 2014, 131, 249–257. [Google Scholar] [CrossRef]
- Solberg, T.R.; Sonesson, A.K.; Woolliams, J.A.; Odegard, J.; Meuwissen, T.H.E. Persistence of accuracy of genome-wide breeding values over generations when including a polygenic effect. Genet. Sel. Evol. 2009, 41, 53. [Google Scholar] [CrossRef]
- Calus, M.P.L.; Veerkamp, R.F. Accuracy of breeding values when using and ignoring the polygenic effect in genomic breeding value estimation with a marker density of one SNP per cM. J. Anim. Breed. Genet. 2007, 124, 362–368. [Google Scholar] [CrossRef]
- Goddard, M. Genomic selection: Prediction of accuracy and maximisation of long term response. Genetica 2009, 136, 245–257. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Bordbar, F.; Jensen, J.; Du, M.; Abied, A.; Guo, W.; Xu, L.; Gao, H.; Zhang, L.; Li, J. Identification and validation of a novel candidate gene regulating net meat weight in Simmental beef cattle based on imputed next-generation sequencing. Cell Prolif. 2020, 53, e12870. [Google Scholar] [CrossRef]
- Roos, A.P.W.D.; Hayes, B.J.; Spelman, R.J.; Goddard, M.E. Linkage Disequilibrium and Persistence of Phase in Holstein-Friesian, Jersey and Angus Cattle. Genetics 2008, 179, 1503–1512. [Google Scholar] [CrossRef]
- Ma, P.; Huang, J.; Gong, W.; Li, X.; Gao, H.; Zhang, Q.; Ding, X.; Wang, C. The impact of genomic relatedness between populations on the genomic estimated breeding values. J. Anim. Sci. Biotechnol. 2018, 9, 64. [Google Scholar] [CrossRef]
- Hill, W.G.; Robertson, A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968, 38, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Vanraden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Wientjes, Y.; Bijma, P.; Vandenplas, J.; Calus, M. Multi-population Genomic Relationships for Estimating Current Genetic Variances within and Genetic Correlations between Populations. Genetics 2017, 207, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, Y.P.; Bowman, P.J.; Goddard, M.E.; Hayes, B.J. A hybrid expectation maximisation and MCMC sampling algorithm to implement Bayesian mixture model based genomic prediction and QTL mapping. BMC Genomics 2016, 17, 744. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, I.; Meuwissen, T.H.E.; Macleod, I.M.; Goddard, M.E. Predicting the effect of reference population on the accuracy of within, across, and multibreed genomic prediction. J. Dairy Sci. 2019, 102, 3155–3174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ma, H.; Li, H.; Xu, L.; Li, H.; Zhu, B.; Hay, E.H.; Xu, L.; Li, J. Multi-trait genomic predictions using GBLUP and Bayesian mixture prior model in beef cattle. Anim. Res. One Health 2023, 1, 17–29. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Pong-Wong, R.; Villanueva, B.; Woolliams, J.A. The impact of genetic architecture on genome-wide evaluation methods. Genetics 2010, 185, 1021–1031. [Google Scholar] [CrossRef]
- Habier, D.; Tetens, J.; Seefried, F.R.; Lichtner, P.; Thaller, G. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 2010, 42, 5. [Google Scholar] [CrossRef]
- Steyn, Y.; Lourenco, D.A.L.; Misztal, I. Genomic predictions in purebreds with a multibreed genomic relationship matrix1. J. Anim. Sci. 2019, 97, 4418–4427. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.C.; Ye, C.J.; Price, A.L.; Zaitlen, N. Transethnic Genetic-Correlation Estimates from Summary Statistics. Am. J. Hum. Genet. 2016, 99, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, C.; Miller, S.; Schenkel, F. Multi-population genomic prediction using a multi-task Bayesian learning model. BMC Genet. 2014, 15, 53. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Technow, F.; Totir, L.R. Using Bayesian Multilevel Whole Genome Regression Models for Partial Pooling of Training Sets in Genomic Prediction. G3 Genes|Genomes|Genetics 2015, 5, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, D.A.L.; Tsuruta, S.; Fragomeni, B.O.; Chen, C.Y.; Herring, W.O.; Misztal, I. Crossbreed evaluations in single-step genomic best linear unbiased predictor using adjusted realized relationship matrices. J. Anim. Sci. 2016, 94, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Makgahlela, M.L.; Strandén, I.; Nielsen, U.S.; Sillanpää, M.J.; Mäntysaari, E.A. The estimation of genomic relationships using breedwise allele frequencies among animals in multibreed populations. J. Dairy Sci. 2013, 96, 5364–5375. [Google Scholar] [CrossRef] [PubMed]
- Makgahlela, M.L.; Strandén, I.; Nielsen, U.S.; Sillanpää, M.J.; Mäntysaari, E.A. Using the unified relationship matrix adjusted by breed-wise allele frequencies in genomic evaluation of a multibreed population. J. Dairy Sci. 2014, 97, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Wientjes, Y.C.; Veerkamp, R.F.; Bijma, P.; Bovenhuis, H.; Schrooten, C.; Calus, M.P. Empirical and deterministic accuracies of across-population genomic prediction. Genet. Sel. Evol. 2015, 47, 5. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.S.; van den Berg, I.; Ma, P.; Brøndum, R.F.; Su, G. Review: How to improve genomic predictions in small dairy cattle populations. Animal 2016, 10, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
| Trait | Number | Mean | SD | C.V | Minimum | Maximum | Heritability |
|---|---|---|---|---|---|---|---|
| 1302 | 544.05 | 80.95 | 14.88 | 318 | 790 | 0.40 (0.04) | |
| 600 | 605.10 | 72.05 | 11.91 | 594 | 1001 | 0.53 (0.06) | |
| 400 | 372 | 36.01 | 9.68 | 295 | 420 | 0.49 (0.07) |
| refPop | Method | SNP Density | |
|---|---|---|---|
| HD | WGS | ||
| HX | GBLUP | 0.41 (0.013) | 0.42 (0.010) |
| BayesR | 0.52 (0.010) | 0.46 (0.008) | |
| WG | GBLUP | 0.34 (0.017) | 0.34 (0.012) |
| BayesR | 0.38 (0.010) | 0.38 (0.011) | |
| YL | GBLUP | 0.28 (0.021) | 0.28 (0.019) |
| BayesR | 0.30 (0.016) | 0.32 (0.011) | |
| refPop | Method | HX | WG | YL | |||
|---|---|---|---|---|---|---|---|
| HD | WGS | HD | WGS | HD | WGS | ||
| HX + WG | -GBLUP | 0.41 (0.014) | 0.44 (0.013) | 0.34 (0.014) | 0.35 (0.015) | 0.10 (0.014) | 0.11 (0.013) |
| -GBLUP | 0.42 (0.015) | 0.46 (0.016) | 0.35 (0.013) | 0.38 (0.013) | 0.10 (0.014) | 0.12 (0.012) | |
| BayesR | 0.48 (0.014) | 0.53 (0.012) | 0.4 (0.016) | 0.44 (0.014) | 0.13 (0.019) | 0.17 (0.016) | |
| HX + YL | -GBLUP | 0.41 (0.015) | 0.44 (0.017) | 0.13 (0.015) | 0.15 (0.013) | 0.3 (0.011) | 0.31 (0.015) |
| -GBLUP | 0.41 (0.019) | 0.45 (0.018) | 0.13 (0.019) | 0.16 (0.015) | 0.32 (0.015) | 0.34 (0.019) | |
| BayesR | 0.43 (0.016) | 0.48 (0.013) | 0.15 (0.014) | 0.19 (0.012) | 0.33 (0.016) | 0.38 (0.011) | |
| WG + YL | -GBLUP | 0.11 (0.018) | 0.12 (0.017) | 0.34 (0.015) | 0.35 (0.018) | 0.31 (0.017) | 0.32 (0.017) |
| -GBLUP | 0.13 (0.017) | 0.15 (0.015) | 0.35 (0.015) | 0.37 (0.017) | 0.33 (0.013) | 0.35 (0.011) | |
| BayesR | 0.18 (0.016) | 0.2 (0.013) | 0.41 (0.018) | 0.44 (0.018) | 0.34 (0.016) | 0.38 (0.014) | |
| HX + WG + YL | -GBLUP | 0.42 (0.013) | 0.44 (0.017) | 0.35 (0.014) | 0.36 (0.016) | 0.31 (0.012) | 0.32 (0.018) |
| -GBLUP | 0.42 (0.019) | 0.45 (0.017) | 0.36 (0.016) | 0.4 (0.013) | 0.32 (0.017) | 0.34 (0.016) | |
| BayesR | 0.45 (0.012) | 0.49 (0.012) | 0.43 (0.018) | 0.48 (0.015) | 0.34 (0.018) | 0.39 (0.013) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.; Li, H.; Ge, F.; Zhao, H.; Zhu, B.; Zhang, L.; Gao, H.; Xu, L.; Li, J.; Wang, Z. Improving Genomic Predictions in Multi-Breed Cattle Populations: A Comparative Analysis of BayesR and GBLUP Models. Genes 2024, 15, 253. https://doi.org/10.3390/genes15020253
Ma H, Li H, Ge F, Zhao H, Zhu B, Zhang L, Gao H, Xu L, Li J, Wang Z. Improving Genomic Predictions in Multi-Breed Cattle Populations: A Comparative Analysis of BayesR and GBLUP Models. Genes. 2024; 15(2):253. https://doi.org/10.3390/genes15020253
Chicago/Turabian StyleMa, Haoran, Hongwei Li, Fei Ge, Huqiong Zhao, Bo Zhu, Lupei Zhang, Huijiang Gao, Lingyang Xu, Junya Li, and Zezhao Wang. 2024. "Improving Genomic Predictions in Multi-Breed Cattle Populations: A Comparative Analysis of BayesR and GBLUP Models" Genes 15, no. 2: 253. https://doi.org/10.3390/genes15020253
APA StyleMa, H., Li, H., Ge, F., Zhao, H., Zhu, B., Zhang, L., Gao, H., Xu, L., Li, J., & Wang, Z. (2024). Improving Genomic Predictions in Multi-Breed Cattle Populations: A Comparative Analysis of BayesR and GBLUP Models. Genes, 15(2), 253. https://doi.org/10.3390/genes15020253





