New Axes of Interaction in Circ_0079593/miR-516b-5p Network in Melanoma Metastasis Cell Lines
Abstract
1. Introduction
2. Results
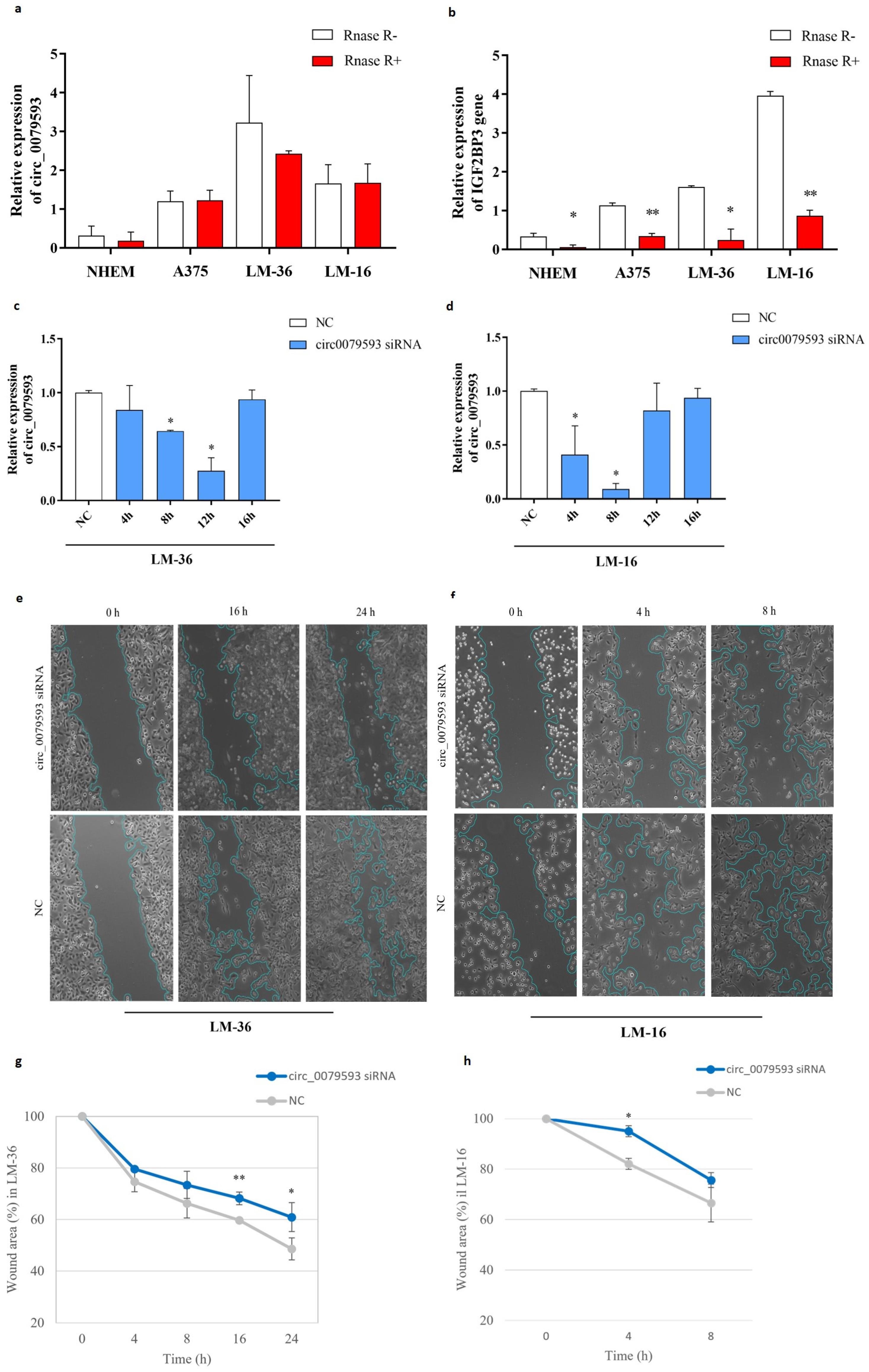
2.1. Expression Pattern of Circ_0079593 in Melanoma Cell Lines and Primary Melanocytic Cells and Its Role in Melanoma Progression
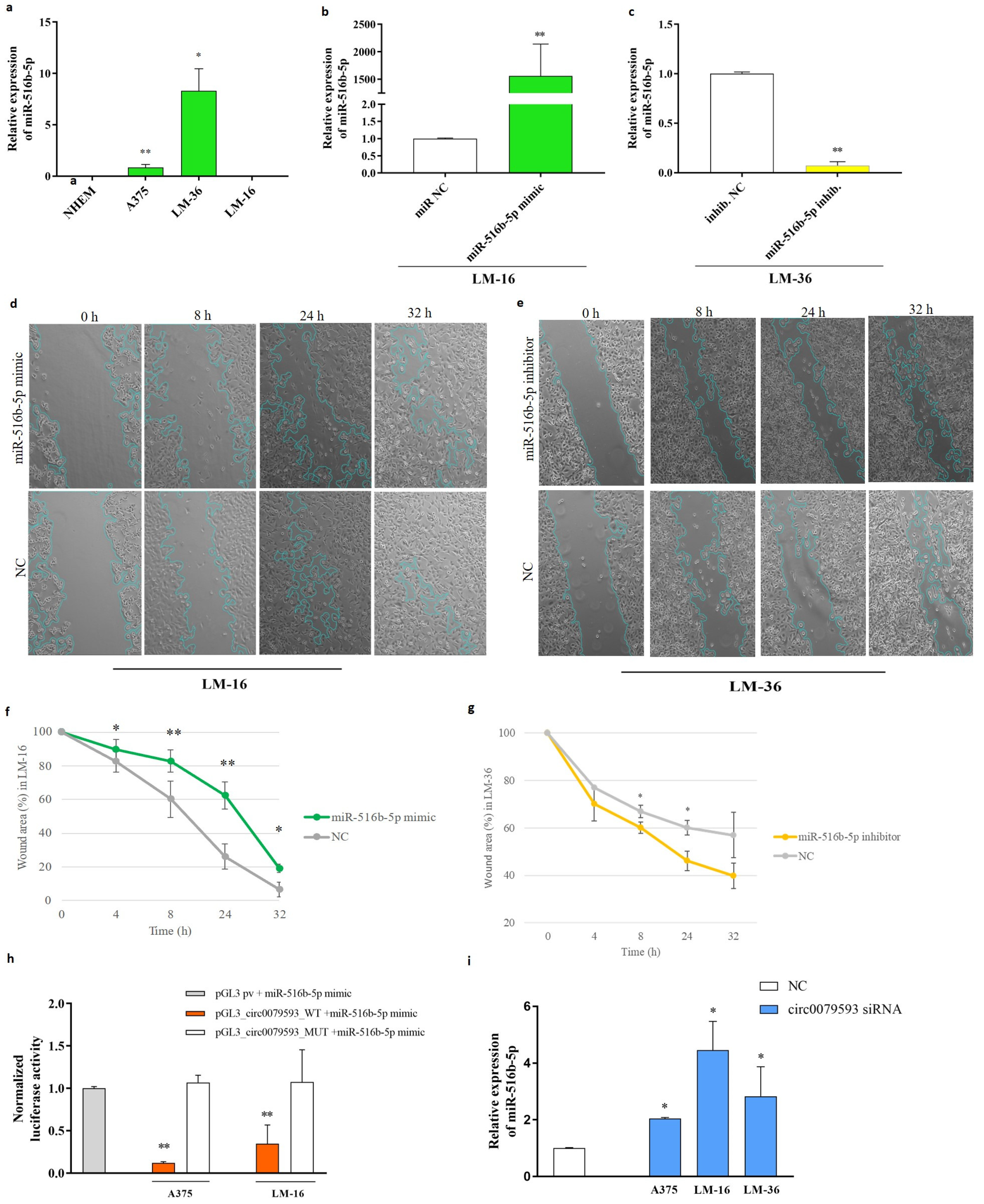
2.2. The Potential of miR-516b-5p as a Target for Circ_0079593 and Its Biological Effects on Melanoma Metastasis
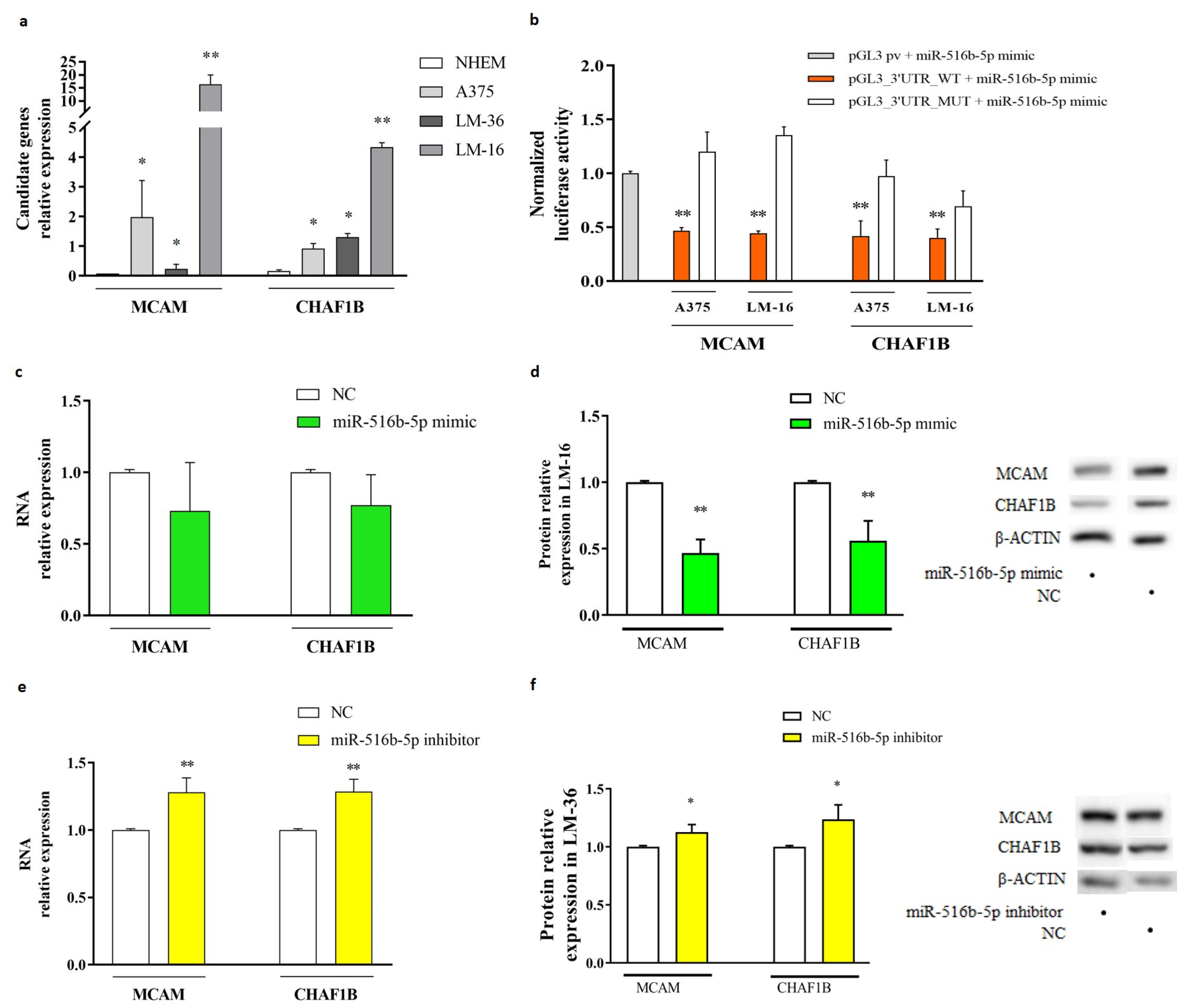
2.3. MiR-516b-5p Targets CHAF1B and MCAM in Melanoma Metastasis Cell Lines
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Transfection Procedures for miRNA and siRNA
4.3. Relative Quantification of miRNA mRNA and CircRNA with Real-Time PCR
4.4. Protein Isolation and Western Blot
4.5. Scratch Wound Healing Test
4.6. Plasmid Construction and Dual Luciferase Reporter Assay
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, Q.; Wang, J. Non-Coding RNAs in Melanoma: Biological Functions and Potential Clinical Applications. Mol. Ther. Oncolytics 2021, 22, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Poniewierska-Baran, A.; Słuczanowska-Głąbowska, S.; Małkowska, P.; Sierawska, O.; Zadroga, Ł.; Pawlik, A.; Niedźwiedzka-Rystwej, P. Role of MiRNA in Melanoma Development and Progression. Int. J. Mol. Sci. 2023, 24, 201. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, J.; Wang, A.; Sun, L.; Qian, L.; Zhou, X.; Liu, Y.; Tang, S.; Chen, X.; Cheng, Y.; et al. Differentially Expressed CircRNAs in Melanocytes and Melanoma Cells and Their Effect on Cell Proliferation and Invasion. Oncol. Rep. 2018, 39, 1813–1824. [Google Scholar] [CrossRef]
- Xin, Z.; Ma, Q.; Ren, S.; Wang, G.; Li, F. The Understanding of Circular RNAs as Special Triggers in Carcinogenesis. Brief. Funct. Genom. 2017, 16, 80–86. [Google Scholar] [CrossRef]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. MiRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Frías-Lasserre, D.; Villagra, C.A. The Importance of NcRNAs as Epigenetic Mechanisms in Phenotypic Variation and Organic Evolution. Front. Microbiol. 2017, 8, 2483. [Google Scholar] [CrossRef]
- Liang, Z.Z.; Guo, C.; Zou, M.M.; Meng, P.; Zhang, T.T. CircRNA-MiRNA-MRNA Regulatory Network in Human Lung Cancer: An Update. Cancer Cell Int. 2020, 20, 173. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long Non-Coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Wan, G.; Liu, Y.; Han, C.; Zhang, X.; Lu, X. Noncoding RNAs in DNA Repair and Genome Integrity. Antioxid. Redox Signal. 2014, 20, 655–677. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Xiao, M.S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wu, Y.; Zeng, B.; Sun, J.; Li, Y.; Luo, J.; Wang, L.; Yi, Z.; Li, H.; Ren, G. CircEHMT1 Inhibits Metastatic Potential of Breast Cancer Cells by Modulating MiR-1233-3p/KLF4/MMP2 Axis. Biochem. Biophys. Res. Commun. 2020, 526, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Mecozzi, N.; Vera, O.; Karreth, F.A. Squaring the Circle: CircRNAs in Melanoma. Oncogene 2021, 40, 5559–5566. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Jara, C.A.C.; Fenske, P.; et al. Loss of a Mammalian Circular RNA Locus Causes MiRNA Deregulation and Affects Brain Function. Science 2017, 357, 1257. [Google Scholar] [CrossRef]
- Horsham, J.L.; Kalinowski, F.C.; Epis, M.R.; Ganda, C.; Brown, R.A.M.; Leedman, P.J. Clinical Potential of MicroRNA-7 in Cancer. J. Clin. Med. 2015, 4, 1668–1687. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Hansen, T.B.; Venø, M.T.; Kjems, J. Circular RNAs in Cancer: Opportunities and Challenges in the Field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef]
- Ruiter, D.J.; Spatz, A.; Van Den Oord, J.J.; Cook, M.G. Pathologic Staging of Melanoma. Semin. Oncol. 2002, 29, 370–381. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, Q. Suppression of Exosomal Hsa_circ_0001005 Eliminates the Vemurafenib Resistance of Melanoma. J. Cancer Res. Clin. Oncol. 2023, 149, 5921–5936. [Google Scholar] [CrossRef] [PubMed]
- Lazăr, A.D.; Dinescu, S.; Costache, M. The Non-Coding Landscape of Cutaneous Malignant Melanoma: A Possible Route to Efficient Targeted Therapy. Cancers 2020, 12, 3378. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Yang, H.; Sun, H.; Hao, M.; Liu, C.; Zheng, Y.; Cong, X.; Jiang, R. Role of Circular RNAs in the Pathogenesis of Malignant Melanoma. Crit. Rev. Eukaryot. Gene Expr. 2022, 32, 21–30. [Google Scholar] [CrossRef] [PubMed]
- HAO, Y.L.; WANG, X.Q. Circ-0079593 Promotes Proliferation and Migration of Melanoma Cells by Sponging MicroRNA-433 and Elevating EGFR Expression. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Jia, Z.; Feng, Y.; Li, Z.; Feng, J. Circular RNA Circ_0079593 Enhances Malignant Melanoma Progression by the Regulation of the MiR-573/ABHD2 Axis. J. Dermatol. Sci. 2021, 102, 7–15. [Google Scholar] [CrossRef]
- Lu, J.; Li, Y. Circ_0079593 Facilitates Proliferation, Metastasis, Glucose Metabolism and Inhibits Apoptosis in Melanoma by Regulating the MiR-516b/GRM3 Axis. Mol. Cell. Biochem. 2020, 475, 227–237. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, K.; Zhou, H.; Wu, Y.; Li, C.; Liu, Y.; Liu, Z.; Xu, Q.; Liu, S.; Xiao, D.; et al. Role of Non-Coding RNAs and RNA Modifiers in Cancer Therapy Resistance. Mol. Cancer 2020, 19, 47. [Google Scholar] [CrossRef]
- Bian, D.; Wu, Y.; Song, G. Novel Circular RNA, Hsa_circ_0025039 Promotes Cell Growth, Invasion and Glucose Metabolism in Malignant Melanoma via the MiR-198/CDK4 Axis. Biomed. Pharmacother. 2018, 108, 165–176. [Google Scholar] [CrossRef]
- Grafanaki, K.; Grammatikakis, I.; Ghosh, A.; Gopalan, V.; Olgun, G.; Liu, H.; Kyriakopoulos, G.C.; Skeparnias, I.; Georgiou, S.; Stathopoulos, C.; et al. Noncoding RNA Circuitry in Melanoma Onset, Plasticity, and Therapeutic Response. Pharmacol. Ther. 2023, 248, 108466. [Google Scholar] [CrossRef]
- Tang, K.; Zhang, H.; Li, Y.; Sun, Q.; Jin, H. Circular RNA as a Potential Biomarker for Melanoma: A Systematic Review. Front. Cell Dev. Biol. 2021, 9, 638548. [Google Scholar] [CrossRef] [PubMed]
- Rapanotti, M.C.; Cugini, E.; Nuccetelli, M.; Terrinoni, A.; Di Raimondo, C.; Lombardo, P.; Costanza, G.; Cosio, T.; Rossi, P.; Orlandi, A.; et al. MCAM/MUC18/CD146 as a Multifaceted Warning Marker of Melanoma Progression in Liquid Biopsy. Int. J. Mol. Sci. 2021, 22, 12416. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Guan, C.W.; Song, Y.; Wang, H. The Multifaceted Role of CD146/MCAM in the Promotion of Melanoma Progression. Cancer Cell Int. 2015, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Mascolo, M.; Vecchione, M.L.; Ilardi, G.; Scalvenzi, M.; Molea, G.; Di Benedetto, M.; Nugnes, L.; Siano, M.; De Rosa, G.; Staibano, S. Overexpression of Chromatin Assembly Factor-1/P60 Helps to Predict the Prognosis of Melanoma Patients. BMC Cancer 2010, 10, 63. [Google Scholar] [CrossRef]
- Reid, A.L.; Millward, M.; Pearce, R.; Lee, M.; Frank, M.H.; Ireland, A.; Monshizadeh, L.; Rai, T.; Heenan, P.; Medic, S.; et al. Markers of Circulating Tumour Cells in the Peripheral Blood of Patients with Melanoma Correlate with Disease Recurrence and Progression. Br. J. Dermatol. 2013, 168, 85–92. [Google Scholar] [CrossRef]
- Pang, Y.; Maxwell, E.; Sindrewicz-Goral, P.; Shapanis, A.; Li, S.; Morgan, M.; Yu, L.G. Galectin-3 Is a Natural Binding Ligand of MCAM (CD146, MUC18) in Melanoma Cells and Their Interaction Promotes Melanoma Progression. Biomolecules 2022, 12, 1451. [Google Scholar] [CrossRef]
- Di, M.; Wang, M.; Miao, J.; Chen, B.; Huang, H.; Lin, C.; Jian, Y.; Li, Y.; Ouyang, Y.; Chen, X.; et al. CHAF1B Induces Radioresistance by Promoting DNA Damage Repair in Nasopharyngeal Carcinoma. Biomed. Pharmacother. 2020, 123, 109748. [Google Scholar] [CrossRef]
- Daniotti, M.; Oggionni, M.; Ranzani, T.; Vallacchi, V.; Campi, V.; Di Stasi, D.; Della Torre, G.; Perrone, F.; Luoni, C.; Suardi, S.; et al. BRAF Alterations Are Associated with Complex Mutational Profiles in Malignant Melanoma. Oncogene 2004, 23, 5968–5977. [Google Scholar] [CrossRef]
- Ye, X.Y.; Xu, L.; Lu, S.; Chen, Z.W. MiR-516a-5p Inhibits the Proliferation of Non-Small Cell Lung Cancer by Targeting HIST3H2A. Int. J. Immunopathol. Pharmacol. 2019, 33, 1–7. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, T.; Wu, C.; Zhang, Y. CircAKT3 Inhibits Glycolysis Balance in Lung Cancer Cells by Regulating MiR-516b-5p/STAT3 to Inhibit Cisplatin Sensitivity. Biotechnol. Lett. 2020, 42, 1123–1135. [Google Scholar] [CrossRef]
- Hanniford, D.; Segura, M.F.; Zhong, J.; Philips, E.; Jirau-Serrano, X.; Darvishian, F.; Berman, R.S.; Shapiro, R.L.; Pavlick, A.C.; Brown, B.; et al. Identification of Metastasis-Suppressive MicroRNAs in Primary Melanoma. J. Natl. Cancer Inst. 2015, 107, 1–13. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Genes | Forward and Reverse Primer | Product |
| CHAF1B | F: 5’-CCTGGAAAAGCCACTCTTGC-3’ | 139 bp |
| R: 5’-ACAGAAGCACGGAATCCTCC-3’ | ||
| MCAM | F: 5’-AAGGAGAGGAAGGTGTGGGT-3’ | 119 bp |
| R: 5’-TGGTCTTGTTCACTTGCCGT-3’ | ||
| TBP | F: 5’-TGTATCCACAGTGAATCTTGG-3’ | 102 bp |
| R: 5’-ATGATTACCGCAGCAAACC-3’ | ||
| IGF2BP3 | F: 5’-CCATAGAAGTTGAGCACTCGGTCC-3’ | 126 bp |
| R: 5’-TCTCCACCACTCCATACAGGACTAG-3’ | ||
| circRNA | Forward and Reverse Primer | Product |
| hsa_circ_0079593 | F: 5’-AATCTGAACGCCTTGGGTCT-3’ | 172 bp |
| R: 5’-CTCAGCTTTGGCACATGTCT-3’ |
| Wild-Type Plasmid | Primers for Vector Construction | Product |
| pGL3_3’UTR_CHAF1B_WT | F: 5’-CCTAAGGTTCTAGAGGAGCGGGACACACTGTAAA-3’ | 404 bp |
| R: 5’-ACCTTAGGTCTAGAAAACAAACACAACTACCTTCCAA-3’ | ||
| pGL3_3’UTR_MCAM_WT | F: 5’-CCTAAGTTCTAGAAATCTGAACGCCTTGGGTCTG-3’ | 418 bp |
| R: 5’-ACCTTAGGTCTAGACTCAGCTTTGGCACATGTCT-3’ | ||
| pGL3_circ_0079593_WT | F: 5’-CCTAAGGTTCTAGAGAGATGGTGGTGGACTGGTC-3’ | 199 bp |
| R: 5’-ACCTTAGGTCTAGAAGTTCCTGGCTTCTGACCAA-3’ | ||
| Mutated Plasmid | Primers Used for Mutagenesis Reaction | |
| pGL3_3’UTR_CHAF1B_MUT | F: 5’-ATACTGAATACAACAGCATCCATATGACTGGAGAAAAATCAGTACACATGTC-3’ | |
| R: 5’-GACATGTGTACTGATTTTTCTCCAGTCATATGGATGCTGTTGTATTCAGTAT-3’ | ||
| pGL3_3’UTR_MCAM_MUT | F: 5’-CAACACTGCAGCTGCAGCTGGATGCTGCTGGG-3’ | |
| R: 5’-CCCAGCAGCATCCAGCTGCAGCTGCAGTGTTG-3’ | ||
| pGL3_circ0079593_MUT | F: 5’-TGCCTTTAACTGTAATAGTGCGCGCTGGATTATACAGCGTCAATTC-3’ | |
| R: 5’-GAATTGACGCTGTATAATCCAGCGCGCACTATTACAGTTAAAGGCA-3’ | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Tomi, E.; Orlandi, E.; Belpinati, F.; Patuzzo, C.; Trabetti, E.; Gomez-Lira, M.; Malerba, G. New Axes of Interaction in Circ_0079593/miR-516b-5p Network in Melanoma Metastasis Cell Lines. Genes 2024, 15, 1647. https://doi.org/10.3390/genes15121647
De Tomi E, Orlandi E, Belpinati F, Patuzzo C, Trabetti E, Gomez-Lira M, Malerba G. New Axes of Interaction in Circ_0079593/miR-516b-5p Network in Melanoma Metastasis Cell Lines. Genes. 2024; 15(12):1647. https://doi.org/10.3390/genes15121647
Chicago/Turabian StyleDe Tomi, Elisa, Elisa Orlandi, Francesca Belpinati, Cristina Patuzzo, Elisabetta Trabetti, Macarena Gomez-Lira, and Giovanni Malerba. 2024. "New Axes of Interaction in Circ_0079593/miR-516b-5p Network in Melanoma Metastasis Cell Lines" Genes 15, no. 12: 1647. https://doi.org/10.3390/genes15121647
APA StyleDe Tomi, E., Orlandi, E., Belpinati, F., Patuzzo, C., Trabetti, E., Gomez-Lira, M., & Malerba, G. (2024). New Axes of Interaction in Circ_0079593/miR-516b-5p Network in Melanoma Metastasis Cell Lines. Genes, 15(12), 1647. https://doi.org/10.3390/genes15121647









