Combined Use of Univariate and Multivariate Approaches to Detect Selection Signatures Associated with Milk or Meat Production in Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Population Stratification
2.3. Selection Signature Detection
2.4. Marker-of-Interest Selection
2.5. Gene and Quantitative Trait Loci Research
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Maiorano, A.M.; Cardoso, D.F.; Carvalheiro, R.; Júnior, G.A.F.; de Albuquerque, L.G.; de Oliveira, H.N. Signatures of selection in Nelore cattle revealed by whole-genome sequencing data. Genomics 2022, 114, 110304. [Google Scholar] [CrossRef] [PubMed]
- Klímová, A.; Kašná, E.; Machová, K.; Brzáková, M.; Přibyl, J.; Vostrý, L. The use of genomic data and imputation methods in dairy cattle breeding. Czech J. Anim. Sci. 2022, 65, 445–453. [Google Scholar] [CrossRef]
- Krivoruchko, A.; Likhovid, A.; Kanibolotskaya, A.; Saprikina, T.; Safaryan, E.; Yatsyk, O. Genome-Wide Search for Associations with Meat Production Parameters in Karachaevsky Sheep Breed Using the Illumina BeadChip 600 K. Genes 2023, 14, 1288. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J.; Meuwissen, T.H. Genomic selection in livestock populations. Genet. Res. 2010, 92, 413–421. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J. Genomic selection. J. Anim. Breed. Genet. 2007, 124, 323–330. [Google Scholar] [CrossRef]
- Kim, E.S.; Elbeltagy, A.R.; Aboul-Naga, A.M.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.M.; Rothschild, M.F. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 2016, 116, 255–264. [Google Scholar] [CrossRef]
- Gouveia, J.J.D.S.; Silva, M.V.G.B.D.; Paiva, S.R.; Oliveira, S.M.P.D. Identification of selection signatures in livestock species. Genet. Mol. Biol. 2014, 37, 330–342. [Google Scholar] [CrossRef]
- Rothammer, S.; Seichter, D.; Förster, M.; Medugorac, I. A genome-wide scan for signatures of differential artificial selection in ten cattle breeds. BMC Genom. 2013, 14, 908. [Google Scholar] [CrossRef]
- Taye, M.; Lee, W.; Jeon, S.; Yoon, J.; Dessie, T.; Hanotte, O.; Mwai, O.A.; Kemp, S.; Cho, S.; Oh, S.J.; et al. Exploring evidence of positive selection signatures in cattle breeds selected for different traits. Mamm. Genome 2017, 28, 528–541. [Google Scholar] [CrossRef]
- Seo, D.; Lee, D.H.; Jin, S.; Won, J.I.; Lim, D.; Park, M.; Kim, T.H.; Lee, H.K.; Kim, S.; Choi, I.; et al. Long-term artificial selection of Hanwoo (Korean) cattle left genetic signatures for the breeding traits and has altered the genomic structure. Sci. Rep. 2022, 12, 6438. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.J.G. Genetic Differentiation among Livestock Breeds—Values for Fst. Animals 2022, 12, 1115. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.H.; Nejati-Javaremi, A.; Moradi-Shahrbabak, M.; Dodds, K.G.; McEwan, J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. 2012, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Moravčíková, N.; Kasarda, R.; Vostrý, L.; Krupová, Z.; Krupa, E.; Lehocká, K.; Olšanská, B.; Trakovická, A.; Nádaský, R.; Židek, R.; et al. Analysis of selection signatures in the beef cattle genome. Czech J. Anim. Sci. 2019, 64, 491–503. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef]
- Dadousis, C.; Veerkamp, R.F.; Heringstad, B.; Pszczola, M.; Calus, M.P. A comparison of principal component regression and genomic REML for genomic prediction across populations. Genet. Sel. 2014, 46, 60. [Google Scholar] [CrossRef]
- Manca, E.; Cesarani, A.; Gaspa, G.; Sorbolini, S.; Macciotta, N.P.; Dimauro, C. Use of the multivariate discriminant analysis for genome-wide association studies in cattle. Animals 2020, 10, 1300. [Google Scholar] [CrossRef]
- Peng, Q.; Zhao, J.; Xue, F. PCA-based bootstrap confidence interval tests for gene-disease association involving multiple SNPs. BMC Genet. 2010, 11, 6. [Google Scholar] [CrossRef]
- Hahs-Vaughn, D.L. Applied Multivariate Statistical Concepts, 1st ed.; Routledge: Oxford, UK; Taylor & Francis: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Dimauro, C.; Nicoloso, L.; Cellesi, M.; Macciotta, N.P.P.; Ciani, E.; Moioli, B.; Pilia, F.; Crepaldi, P. Selection of discriminant SNP markers for breed and geographic assignment of Italian sheep. Small Rumin. Res. 2015, 128, 27–33. [Google Scholar] [CrossRef]
- Duforet-Frebourg, N.; Luu, K.; Laval, G.; Bazin, E.; Blum, M.G. Detecting genomic signatures of natural selection with principal component analysis: Application to the 1000 genomes data. Mol. Biol. Evol. 2016, 33, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Sorbolini, S.; Gaspa, G.; Steri, R.; Dimauro, C.; Cellesi, M.; Stella, A.; Marras, G.; Marsan, P.A.; Valentini, A.; Macciotta, N.P.P. Use of canonical discriminant analysis to study signatures of selection in cattle. Genet. Sel. 2016, 48, 58. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Li, W.; Li, F.; Yue, X. Selection signature analysis reveals genes underlying sheep milking performance. Arch. Anim. Breed. 2019, 62, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, H.; Xie, J.; Wang, Z.; Ma, Y. Trait-specific selection signature detection reveals novel loci of meat quality in large white pigs. Front. Genet. 2021, 12, 761252. [Google Scholar] [CrossRef]
- Paim, T.D.P.; dos Santos, C.A.; de Faria, D.A.; Paiva, S.R.; McManus, C. Genomic selection signatures in Brazilian sheep breeds reared in a tropical environment. Livest. Sci. 2022, 258, 104865. [Google Scholar] [CrossRef]
- de Villemereuil, P.; Frichot, É.; Bazin, É.; François, O.; Gaggiotti, O.E. Genome scan methods against more complex models: When and how much should we trust them? Mol. Ecol. 2014, 23, 2006–2019. [Google Scholar] [CrossRef]
- Cesarani, A.; Sorbolini, S.; Criscione, A.; Bordonaro, S.; Pulina, G.; Battacone, G.; Marletta, D.; Gaspa, G.; Macciotta, N.P.P. Geno-me-wide variability and selection signatures in Italian island cattle breeds. Anim. Genet. 2018, 49, 371–383. [Google Scholar] [CrossRef]
- Saravanan, K.A.; Panigrahi, M.; Kumar, H.; Bhushan, B.; Dutt, T.; Mishra, B.P. Selection signatures in livestock genome: A review of concepts, approaches and applications. Livest. Sci. 2020, 241, 104257. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Ben-Jemaa, S.; Perini, F.; Cendron, F.; Biscarini, F.; Lasagna, E.; Penasa, M.; Cassandro, M. Genome-wide mapping of signatures of selection using a high-density array identified candidate genes for growth traits and local adaptation in chickens. Genet. Sel. 2023, 55, 20. [Google Scholar] [CrossRef]
- Sempéré, G.; Moazami-Goudarzi, K.; Eggen, A.; Laloë, D.; Gautier, M.; Flori, L. WIDDE: A Web-Interfaced next generation database for genetic diversity exploration, with a first application in cattle. BMC Genet. 2015, 16, 940. [Google Scholar] [CrossRef]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connnell, J.; Moore, S.S.; Smith, T.P.L.; Sonstegard, T.S.; et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef] [PubMed]
- Gautier, M.; Laloë, D.; Moazami-Goudarzi, K. Insights into the genetic history of French cattle from dense SNP data on 47 worldwide breeds. PLoS ONE 2010, 5, e13038. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 1 August 2024).
- Pintus, E.; Sorbolini, S.; Albera, A.; Gaspa, G.; Dimauro, C.; Steri, R.; Marras, G.; Macciotta, N.P.P. Use of locally weighted scatterplot smoothing (lowess) regression to study selection signatures in piedmontese and italian brown cattle breeds. Anim. Genet. 2014, 45, 1–11. [Google Scholar] [CrossRef]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D.L. The mahalanobis distance. Chemom. Intell. Lab. Syst. 2000, 50, 1–18. [Google Scholar] [CrossRef]
- Fonseca, P.A.; Suárez-Vega, A.; Marras, G.; Cánovas, A. Genomic Annotation in Livestock for positional candidate LOci: GALLO. Dim (QTLmark.) 2020, 1, 7. Available online: https://cran.uni-muenster.de/web/packages/GALLO/vignettes/GALLO.html (accessed on 29 October 2024).
- Di Stasio, L.; Albera, A.; Pauciullo, A.; Cesarani, A.; Macciotta, N.P.P.; Gaspa, G. Genetics of Arthrogryposis and Macroglossia in Piemontese Cattle Breed. Animals 2020, 10, 1732. [Google Scholar] [CrossRef]
- Maiorano, A.M.; Lourenco, D.L.; Tsuruta, S.; Ospina, A.M.T.; Stafuzza, N.B.; Masuda, Y.; Filho, A.E.V.; do Santos Goncalves Cyrillo, J.N.; Curi, R.A.; Silva, J.A.I.D.V. Assessing genetic architecture and signatures of selection of dual purpose Gir cattle populations using genomic information. PLoS ONE 2018, 13, e0200694. [Google Scholar] [CrossRef]
- Sharma, A.; Lee, S.H.; Lim, D.; Chai, H.H.; Choi, B.H.; Cho, Y. A genome-wide assessment of genetic diversity and population structure of Korean native cattle breeds. BMC Genet. 2016, 17, 139. [Google Scholar] [CrossRef]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.S.; Oget, C.; Palhière, I.; Crisà, A.; Catillo, G.; Steri, R.; et al. AdaptMap Consortium. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. 2018, 50, 57. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, A.A.; Deniskova, T.E.; Yudin, N.S.; Dotsev, A.V.; Khamiruev, T.N.; Selionova, M.I.; Egorov, S.V.; Reyer, H.; Wimmers, K.; Brem, G.; et al. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia. BMC Genom. 2019, 20, 294. [Google Scholar] [CrossRef] [PubMed]
- Toro-Ospina, A.M.; Herrera Rios, A.C.; Bizarria Santos, W.; Pimenta Schettini, G.; Vallejo Aristizabal, V.H.; Tovar Claros, G.; Morea, E.G.O. Genetic architecture and signatures of selection in the Caqueteño creole (Colombian native cattle). Diversity 2024, 14, 828. [Google Scholar] [CrossRef]
- de Siqueira e Oliveira, F.S.; Giana, H.E.; Silveira Jr, L. Discrimination of selected species of pathogenic bacteria using near-infrared Raman spectroscopy and principal components analysis. J. Biomed. Opt. 2012, 17, 107004. [Google Scholar] [CrossRef]
- Kasarda, R.; Moravčíková, N.; Mészáros, G.; Simčič, M.; Zaborski, D. Classification of cattle breeds based on the random forest approach. Livest. Sci. 2023, 267, 105143. [Google Scholar] [CrossRef]
- Lee, S.; Chung, H.; Choi, H.; Cha, K. Using combinations of principal component scores from different spectral ranges in near-infrared region to improve discrimination for samples of complex composition. Microchem. J. 2010, 95, 96–101. [Google Scholar] [CrossRef]
- Cruz-Castillo, J.G.; Ganeshanandam, S.; MacKay, B.R.; Lawes, G.S.; Lawoko, C.R.O.; Woolley, D.J. Applications of canonical discriminant analysis in horticultural. research. HortScience 1994, 29, 1115–1119. [Google Scholar] [CrossRef]
- Strauss, R.E. Discriminating groups of organisms. Morphometr. Nonmorphometr. 2010, 124, 73–91. [Google Scholar] [CrossRef]
- Dimauro, C.; Cellesi, M.; Steri, R.; Gaspa, G.; Sorbolini, S.; Stella, A.; Macciotta, N.P.P. Use of the canonical discriminant analysis to select SNP markers for bovine breed assignment and traceability purposes. Anim. Genet. 2013, 44, 377–382. [Google Scholar] [CrossRef]
- Berry, D.P. Invited review: Beef-on-dairy—The generation of crossbred beef × dairy cattle. J. Dairy Sci. 2021, 104, 3789–3819. [Google Scholar] [CrossRef]
- Esrafili Taze Kand Mohammaddiyeh, M.; Rafat, S.A.; Shodja, J.; Javanmard, A.; Esfandyari, H. Selective genotyping to implement genomic selection in beef cattle breeding. Front. Genet. 2023, 14, 1083106. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, M. Udder Health and Milk Composition, with Special Reference to Beef Cows. A Literature Review; Swedish University of Agricultural Sciences: Skara, Sweden, 2020; pp. 1–52. [Google Scholar]
- Rutledge, J.J.; Robison, O.W.; Ahlschwede, W.T.; Legates, J.E. Milk yield and its influence on 205-day weight of beef calves. J. Anim. Sci. 1971, 33, 563–567. [Google Scholar] [CrossRef]
- McHugh, N.; Cromie, A.R.; Evans, R.D.; Berry, D.P. Validation of national genetic evaluations for maternal beef cattle traits using Irish field data. J. Anim. Sci. 2014, 92, 1423–1432. [Google Scholar] [CrossRef]
- Pires, B.C.; Tholon, P.; Buzanskas, M.E.; Sbardella, A.P.; Rosa, J.O.; da Silva, L.O.C.; de Almeida Torres Jùnior, R.A.; Munari, D.P.; de Alencar, M.M. Genetic analyses on bodyweight, reproductive, and carcass traits in composite beef cattle. Anim. Prod. Sci. 2016, 57, 415–421. [Google Scholar] [CrossRef]
- Belk, K.E.; George, M.H.; Tatum, J.D.; Hilton, G.G.; Miller, R.K.; Koohmaraie, M.; Reagan, J.O.; Smith, G.C. Evaluation of the Tendertec beef grading instrument to predict the tenderness of steaks from beef carcasses. J. Anim. Sci. 2001, 79, 688–697. [Google Scholar] [CrossRef]
- Culioli, J. Meat tenderness: Mechanical assessment. In Expression of Tissue Proteinases and Regulation of Protein Degradation as Related to Meat Quality; Ouali, A., DeMeyer, D.I., Smulders, F.J.M., Eds.; ECCEAMST: Utrecht, The Netherlands, 1995; pp. 239–263. [Google Scholar]
- Silva, D.R.; Torres Filho, R.A.; Cazedey, H.P.; Fontes, P.R.; Ramos, A.L.; Ramos, E.M. Comparison of Warner–Bratzler shear force values between round and square cross-section cores from cooked beef and pork Longissimus muscle. Meat Sci. 2015, 103, 1–6. [Google Scholar] [CrossRef]
- Maltin, C.; Balcerzak, D.; Tilley, R.; Delday, M. Determinants of meat quality: Tenderness. Proc. Nutr. Soc. 2003, 62, 337–347. [Google Scholar] [CrossRef]
- Taye, M.; Yoon, J.; Dessie, T.; Cho, S.; Oh, S.J.; Lee, H.K.; Kim, H. Deciphering signature of selection affecting beef quality traits in Angus cattle. Genes Genom. 2018, 40, 63–75. [Google Scholar] [CrossRef]
- Berry, D.P.; Conroy, S.; Pabiou, T.; Cromie, A.R. Animal breeding strategies can improve meat quality attributes within entire populations. Meat Sci. 2017, 132, 6–18. [Google Scholar] [CrossRef]
- Mateescu, R.G.; Garrick, D.J.; Garmyn, A.J.; VanOverbeke, D.L.; Mafi, G.G.; Reecy, J.M. Genetic parameters for sensory traits in longissimus muscle and their associations with tenderness, marbling score, and intramuscular fat in Angus cattle. J. Anim. Sci. 2015, 93, 21–27. [Google Scholar] [CrossRef]
- Minick, J.A.; Dikeman, M.E.; Pollak, E.J.; Wilson, D.E. Heritability and correlation estimates of Warner-Bratzler shear force and carcass traits from Angus-, Charolais-, Hereford-, and Simmental-sired cattle. Can. J. Anim. Sci. 2004, 84, 599–609. [Google Scholar] [CrossRef]
- Moravčikova, N.; Trakovicka, A.; Kadlečik, O.; Kasarda, R. Genomic signatures of selection in cattle through variation of allele frequencies and linkage disequilibrium. J. Cent. Eur. Agric. 2019, 20, 576–580. [Google Scholar] [CrossRef]
- Hu, H.; Mu, T.; Ma, Y.; Wang, X.; Ma, Y. Analysis of longevity traits in Holstein cattle: A review. Front. Genet. 2021, 12, 695543. [Google Scholar] [CrossRef] [PubMed]
- Essl, A. Longevity in dairy cattle breeding: A review. Livest. Prod. Sci. 1998, 57, 79–89. [Google Scholar] [CrossRef]
- Olechnowicz, J.; Kneblewski, P.; Jaśkowski, J.M.; Włodarek, J. Effect of selected factors on longevity in cattle: A review. J. Anim. Plant Sci. 2016, 26, 1533–1541. [Google Scholar]
- De Vries, A.; Marcondes, M.I. Overview of factors affecting productive lifespan of dairy cows. Animal 2020, 14 (Suppl. S1), s155–s164. [Google Scholar] [CrossRef]
- Rauw, W.M.; Kanis, E.; Noordhuizen-Stassen, E.N.; Grommers, F.J. Undesirable side effects of selection for high production efficiency in farm animals: A review. Livest. Prod. Sci. 1998, 56, 15–33. [Google Scholar] [CrossRef]
- Purfield, D.C.; Bradley, D.G.; Evans, R.D.; Kearney, F.J.; Berry, D.P. Genome-wide association study for calving performance using high-density genotypes in dairy and beef cattle. Genet. Sel. 2015, 47, 47. [Google Scholar] [CrossRef]
- Mao, X.; Kadri, N.K.; Thomasen, J.R.; De Koning, D.J.; Sahana, G.; Guldbrandtsen, B. Fine mapping of a calving QTL on Bos taurus autosome 18 in Holstein cattle. J. Anim. Breed. Genet. 2016, 133, 207–218. [Google Scholar] [CrossRef]
- Sigdel, A.; Bisinotto, R.S.; Peñagaricano, F. Genes and pathways associated with pregnancy loss in dairy cattle. Sci. Rep. 2021, 11, 13329. [Google Scholar] [CrossRef]
- Dessie, S.W.; Rings, F.; Holker, M.; Gilles, M.; Jennen, D.; Tholen, E.; Havlicek, V.; Besenfelder, U.; Sukhorukov, V.L.; Zimmermann, U.; et al. Dielectrophoretic behavior of in vitro-derived bovine metaphase II oocytes and zygotes and its relation to in vitro embryonic developmental competence and mRNA expression pattern. Reproduction 2007, 133, 931–946. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bilotto, S.; Boni, R.; Russo, G.L.; Lioi, M.B. Meiosis progression and donor age affect expression profile of DNA repair genes in bovine oocytes. Zygote 2015, 23, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, D.; Worku, D.; Rings, F.; Phatsara, C.; Tholen, E.; Schellander, K.; Hoelker, M. Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Mol. Reprod. Dev. 2009, 76, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.; Villa, R.; Villa, C.; Gonzalez, V.; Montano, M.; Medina, G.; Mahadevan, P. Inspection of real and imputed genotypes reveled 76 SNPs associated to rear udder height in Holstein cattle. J. Adv. Vet. Anim. Res. 2020, 7, 234. [Google Scholar] [CrossRef]
- Pereira, G.; Guo, Y.; Silva, E.; Bevilacqua, C.; Charpigny, G.; Lopes-da-Costa, L.; Humblot, P. Progesterone differentially affects the transcriptomic profiles of cow endometrial cell types. BMC Genom. 2022, 23, 82. [Google Scholar] [CrossRef]
- Raschia, M.A.; Maizon, D.O.; Amadio, A.F.; Nani, J.P.; Poli, M.A. Quantitative trait loci exploration and characterization of gestation length in Holstein cattle. Theriogenology 2024, 215, 43–49. [Google Scholar] [CrossRef]
- Yuan, G.; Bai, F.; Zhang, Q.; An, X.; Wang, Z.; Lei, C.; Dang, R. Dynamic transcriptome profiles and novel markers in bovine spermatogenesis revealed by single-cell sequencing. J. Integr. Agric. 2023, 23, 2362–2378. [Google Scholar] [CrossRef]
- Blanco, F.C.; Soria, M.A.; Klepp, L.I.; Bigi, F. ERAP1 and PDE8A are downregulated in cattle protected against bovine tuberculosis. J. Mol. Microbiol. Biotechnol. 2017, 27, 237–245. [Google Scholar] [CrossRef]
- Nayak, S.S.; Panigrahi, M.; Rajawat, D.; Ghildiyal, K.; Sharma, A.; Parida, S.; Bhushan, B.; Dutt, T. Comprehensive selection signature analyses in dairy cattle exploiting purebred and crossbred genomic data. Mamm. Genome 2023, 34, 615–631. [Google Scholar] [CrossRef]
- Ayalew, W.; Wu, X.Y.; Tarekegn, G.M.; Min, C.H.U.; Liang, C.N.; Tessema, T.S.; Ping, Y.A.N. Signatures of positive selection for local adaptation of African Native Cattle populations: A review. J. Integr. Agric. 2023, 22, 1967–1984. [Google Scholar] [CrossRef]
- Flori, L.; Fritz, S.; Jaffrézic, F.; Boussaha, M.; Gut, I.; Heath, S.; Foulley, J.L.; Gautier, M. The genome response to artificial selection: A case study in dairy cattle. PLoS ONE 2009, 4, e6595. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, D.M.; Craig, A.W.; Senis, Y.A.; Greer, P.A. Absence of Fer protein-tyrosine kinase exacerbates leukocyte recruitment in response to endotoxin. J. Immunol. 2002, 168, 4930–4935. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, P.; Singaravadivelan, A.; Mishra, A.; Jagadeesan, K.; Bakyaraj, S.; Suresh, R.; Sivakumar, T. Whole-genome comparative analysis reveals genetic mechanisms of disease resistance and heat tolerance of tropical Bos indicus cattle breeds. Genome 2022, 64, 241–254. [Google Scholar] [CrossRef]
- Connor, E.E.; Siferd, S.; Elsasser, T.H.; Evock-Clover, C.M.; Van Tassell, C.P.; Sonstegard, T.S.; Fernandes, V.M.; Capuco, A.V. Effects of increased milking frequency on gene expression in the bovine mammary gland. BMC Genom. 2008, 9, 362. [Google Scholar] [CrossRef]
- Halli, K.; Vanvanhossou, S.F.; Bohlouli, M.; König, S.; Yin, T. Identification of candidate genes on the basis of SNP by time-lagged heat stress interactions for milk production traits in German Holstein cattle. PLoS ONE 2021, 16, e0258216. [Google Scholar] [CrossRef]
- Núñez Soto, S.G.; Berruecos Villalobos, J.M.; Moreno, N.C.; Magaña-Monforte, J.G.; Ochoa-Galván, P.; Ulloa-Arvizu, R.; Toledo-Alvarado, H.O. Genome-wide association study for heat stress resistance in Brown Swiss cattle in Yucatan, Mexico. Vet. Mex. 2023, 10, 1–13. [Google Scholar] [CrossRef]
- Ben-Jemaa, S.; Mastrangelo, S.; Lee, S.H.; Lee, J.H.; Boussaha, M. Genome-wide scan for selection signatures reveals novel insights into the adaptive capacity in local North African cattle. Sci. Rep. 2020, 10, 19466. [Google Scholar] [CrossRef]
- Kiser, J.N.; Cornmesser, M.A.; Blackburn, R.; McGuirk, S.M.; Taylor, J.F.; Seabury, C.M.; The Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team; Neibergs, H.L. Validating loci associated with bovine respiratory disease complex in pre-weaned Holstein calves. Anim. Genet. 2020, 51, 91–94. [Google Scholar] [CrossRef]
- Bhat, S.A.; Elnaggar, M.; Hall, T.J.; McHugo, G.P.; Reid, C.; MacHugh, D.E.; Meade, K.G. Preferential differential gene expression within the WC1. 1+ γδ T cell compartment in cattle naturally infected with Mycobacterium bovis. Front. Immunol. 2023, 14, 1265038. [Google Scholar] [CrossRef]
- Kieckens, E.; Rybarczyk, J.; Li, R.W.; Vanrompay, D.; Cox, E. Potential immunosuppressive effects of Escherichia coli O157: H7 experimental infection on the bovine host. BMC Genom. 2016, 17, 1049. [Google Scholar] [CrossRef]
- Lindholm-Perry, A.K.; Freetly, H.C.; Oliver, W.T.; Rempel, L.A.; Keel, B.N. Genes associated with body weight gain and feed intake identified by meta-analysis of the mesenteric fat from crossbred beef steers. PLoS ONE 2020, 15, e0227154. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Bahbahani, H.; Moioli, B.; Ahbara, A.; Al Abri, M.; Almathen, F.; da Silva, A.; Belabdi, I.; Portolano, B.; Mwacharo, J.M.; et al. Novel and known signals of selection for fat deposition in domestic sheep breeds from Africa and Eurasia. PLoS ONE 2019, 14, e0209632. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Gil, B.; Arranz, J.J.; Wiener, P. An interpretive review of selective sweep studies in Bos taurus cattle populations: Identification of unique and shared selection signals across breeds. Front. Gen. 2015, 6, 167. [Google Scholar] [CrossRef]
- Hocquette, J.F.; Lehnert, S.; Barendse, W.; Cassar-Malek, I.; Picard, B. Recent advances in cattle functional genomics and their application to beef quality. Animal 2007, 1, 159–173. [Google Scholar] [CrossRef]
- Li, B.; Fang, L.; Null, D.J.; Hutchison, J.L.; Connor, E.E.; VanRaden, P.M.; VandeHaar, M.J.; Tempelman, R.J.; Weigel, K.A.; Cole, J.B. High-density genome-wide association study for residual feed intake in Holstein dairy cattle. J. Dairy Sci. 2019, 102, 11067–11080. [Google Scholar] [CrossRef]
- Hardie, L.C.; VandeHaar, M.J.; Tempelman, R.J.; Weigel, K.A.; Armentano, L.E.; Wiggans, G.R.; Veerkamp, R.F.; de Haas, Y.; Coffey, M.P.; Connor, E.E.; et al. The genetic and biological basis of feed efficiency in mid-lactation Holstein dairy cows. J. Dairy Sci. 2017, 100, 9061–9075. [Google Scholar] [CrossRef]
- Al-Husseini, W.; Chen, Y.; Gondro, C.; Herd, R.M.; Gibson, J.P.; Arthur, P.F. Characterization and profiling of liver microRNAs by RNA-sequencing in cattle divergently selected for residual feed intake. Asian-Australas. J. Anim. Sci. 2016, 29, 1371. [Google Scholar] [CrossRef]
- Vargas, G.; Neves, H.H.; Camargo, G.M.F.; Cardoso, V.; Munari, D.P.; Carvalheiro, R. Genome-wide association study and functional analysis of feet and leg conformation traits in Nellore cattle. J. Anim. Sci. 2018, 96, 1617–1627. [Google Scholar] [CrossRef]
- Atashi, H.; Chen, Y.; Wilmot, H.; Vanderick, S.; Hubin, X.; Soyeurt, H.; Gengler, N. Single-step genome-wide association for selected milk fatty acids in Dual-Purpose Belgian Blue cows. J. Dairy Sci. 2023, 106, 6299–6315. [Google Scholar] [CrossRef]
- Carvalheira, J.; Salem, M.M.I.; Thompson, G.; Chen, S.Y.; Beja-Pereira, A. Genome-wide association study for milk and protein yields in Portuguese Holstein cattle. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014; Volume 131, pp. 131–183. [Google Scholar]
- Chen, J.; Zhong, J.; He, X.; Li, X.; Ni, P.; Safner, T.; Šprem, N.; Han, J. The de novo assembly of a European wild boar genome revealed unique patterns of chromosomal structural variations and segmental duplications. Anim. Genet. 2022, 53, 281–292. [Google Scholar] [CrossRef]
- Bekele, R.; Taye, M.; Abebe, G.; Meseret, S. Genomic regions and candidate genes associated with milk production traits in Holstein and its crossbred cattle: A review. Int. J. Genom. 2023, 2023, 8497453. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhang, C.; Song, W.; Zhao, X.; Cheng, Y.; Liu, J.; Su, J. Single-cell RNA-seq and single-cell bisulfite-sequencing reveal insights into yak preimplantation embryogenesis. J. Biol. Chem. 2024, 300, 105562. [Google Scholar] [CrossRef] [PubMed]
- Hasankhani, A.; Bahrami, A.; Mackie, S.; Maghsoodi, S.; Alawamleh, H.S.K.; Sheybani, N.; Dehkordi, F.S.; Rajabi, F.; Javanmard, G.; Khadem, H.; et al. In-depth systems biological evaluation of bovine alveolar macrophages suggests novel insights into molecular mechanisms underlying Mycobacterium bovis infection. Front. Microbiol. 2022, 13, 1041314. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.; Zhang, S.; Wang, S.; Wu, X.; Zhang, Q.; Liu, L.; Li, Y.; Qiao, L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS ONE 2014, 9, e96186. [Google Scholar] [CrossRef] [PubMed]
- Wathes, D.C.; Cheng, Z.; Salavati, M.; Buggiotti, L.; Takeda, H.; Tang, L.; Becker, F.; Ingvartsen, K.I.; Ferris, C.; Hostens, M.; et al. Relationships between metabolic profiles and gene expression in liver and leukocytes of dairy cows in early lactation. J. Dairy Sci. 2021, 104, 3596–3616. [Google Scholar] [CrossRef]
- Illa, S.K.; Mumtaz, S.; Nath, S.; Mukherjee, S.; Mukherjee, A. Characterization of runs of Homozygosity revealed genomic inbreeding and patterns of selection in indigenous sahiwal cattle. J. Appl. Genet. 2024, 65, 167–180. [Google Scholar] [CrossRef]
- Sassi, N.B.; González-Recio, Ó.; de Paz-Del Río, R.; Rodríguez-Ramilo, S.T.; Fernández, A.I. Associated effects of copy number variants on economically important traits in Spanish Holstein dairy cattle. J. Dairy Sci. 2016, 99, 6371–6380. [Google Scholar] [CrossRef]
- Mao, C.; Ju, X.; Cheng, H.; Huang, X.; Jiang, F.; Yao, Y.; Lan, X.; Song, E. Determination of genetic variation within the DYRK2 gene and its associations with milk traits in cattle. Arch. Anim. Breed. 2020, 63, 315–323. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, J.; Zhang, P.; Huang, X.; Yang, W.; Liu, R.; Sun, Q.; Lu, Y.; Zhang, M.; Fu, Q. Single-cell RNA sequencing uncovers dynamic roadmap and cell-cell communication during buffalo spermatogenesis. iScience 2023, 26, 105733. [Google Scholar] [CrossRef]
- Riley, D.G.; Welsh Jr, T.H.; Gill, C.A.; Hulsman, L.L.; Herring, A.D.; Riggs, P.K.; Sawyer, J.E.; Sanders, J.O. Whole genome association of SNP with newborn calf cannon bone length. Livest. Sci. 2013, 155, 186–196. [Google Scholar] [CrossRef]
- Baba, T.; Morota, G.; Kawakami, J.; Gotoh, Y.; Oka, T.; Masuda, Y.; Brito, L.F.; Cockrum, R.R.; Kawahara, T. Longitudinal genome-wide association analysis using a single-step random regression model for height in Japanese Holstein cattle. JDS Commun. 2023, 4, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Purfield, D.C.; Evans, R.D.; Carthy, T.R.; Berry, D.P. Genomic regions associated with gestation length detected using whole-genome sequence data differ between dairy and beef cattle. Front. Genet. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Jiang, J.; Li, B.; Zhou, Y.; Freebern, E.; Vanraden, P.M.; Cole, J.B.; Liu, G.E.; Ma, L. Genetic and epigenetic architecture of paternal origin contribute to gestation length in cattle. Commun. Biol. 2019, 2, 100. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Tan, X.; Jin, Q.; Zhan, W.; Liu, G.; Cui, X.; Wang, J.; Meng, X.; Zhu, R.; Wang, K. Multiomics analyses of Jining Grey goat and Boer goat reveal genomic regions associated with fatty acid and amino acid metabolism and muscle development. Anim. Biosci. 2024, 37, 982. [Google Scholar] [CrossRef] [PubMed]
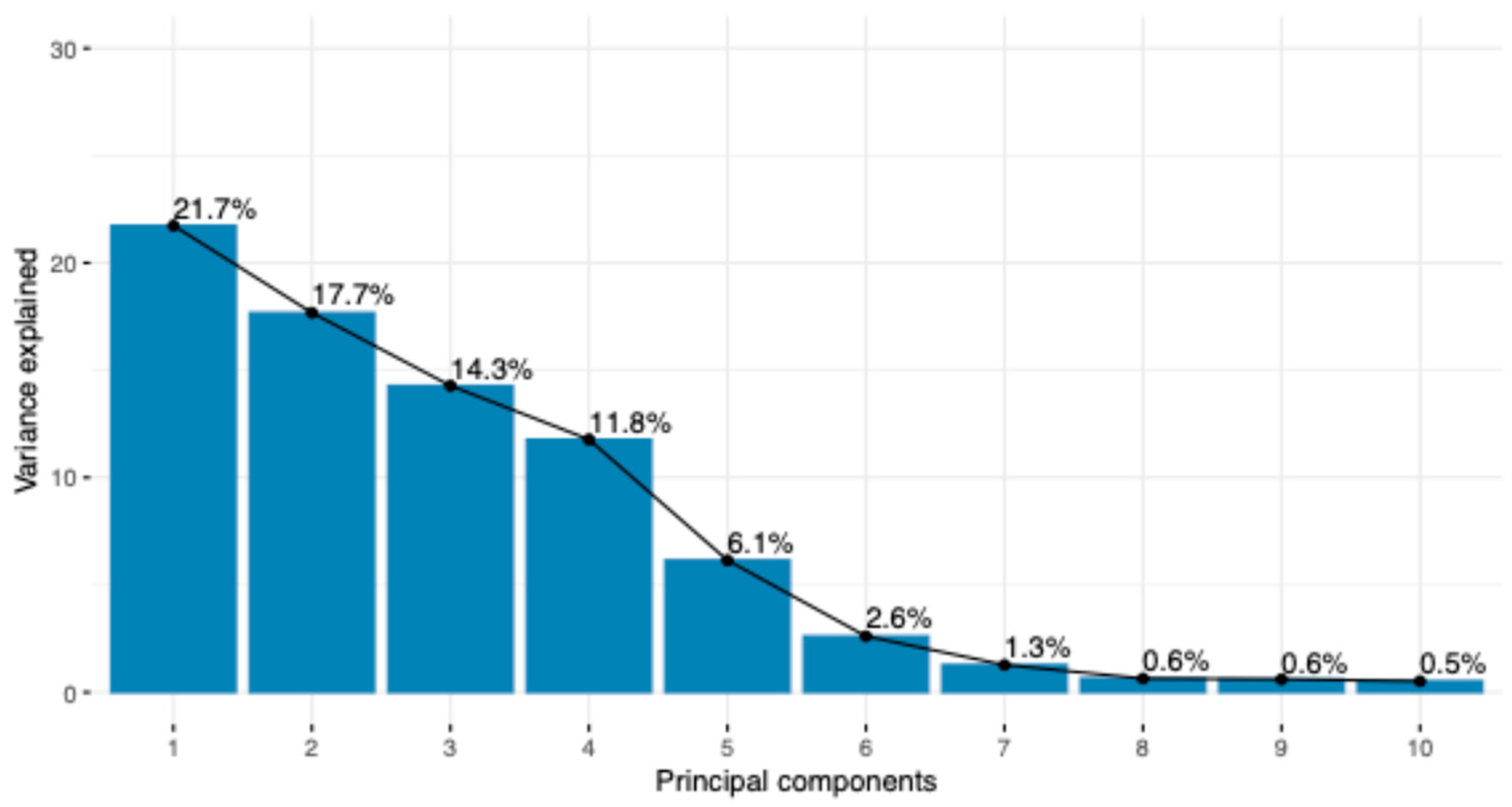

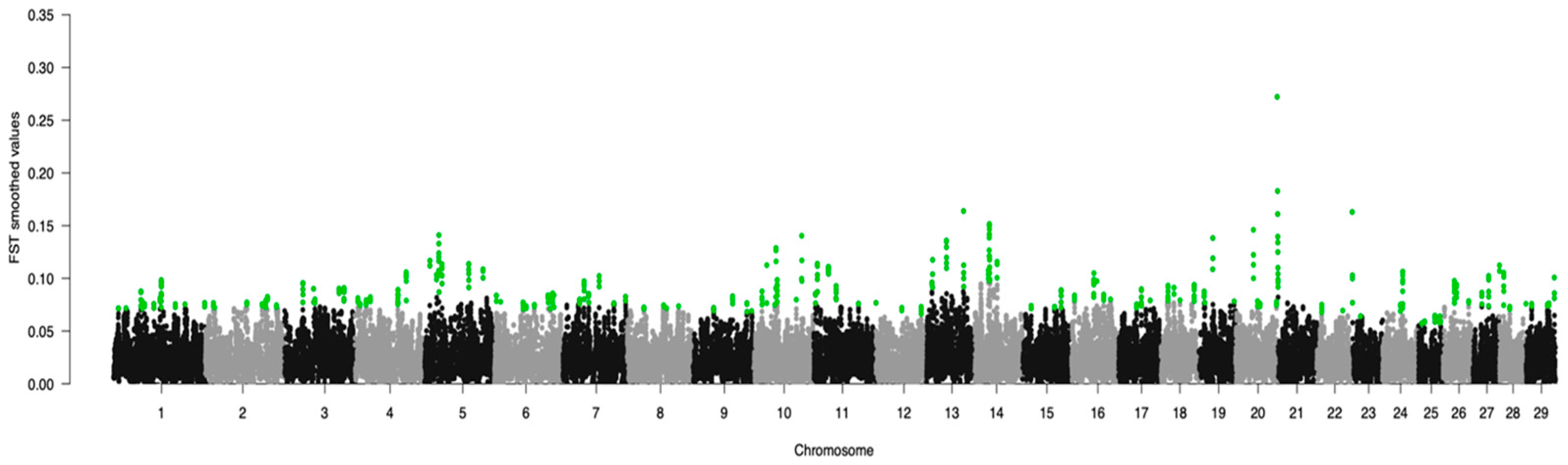

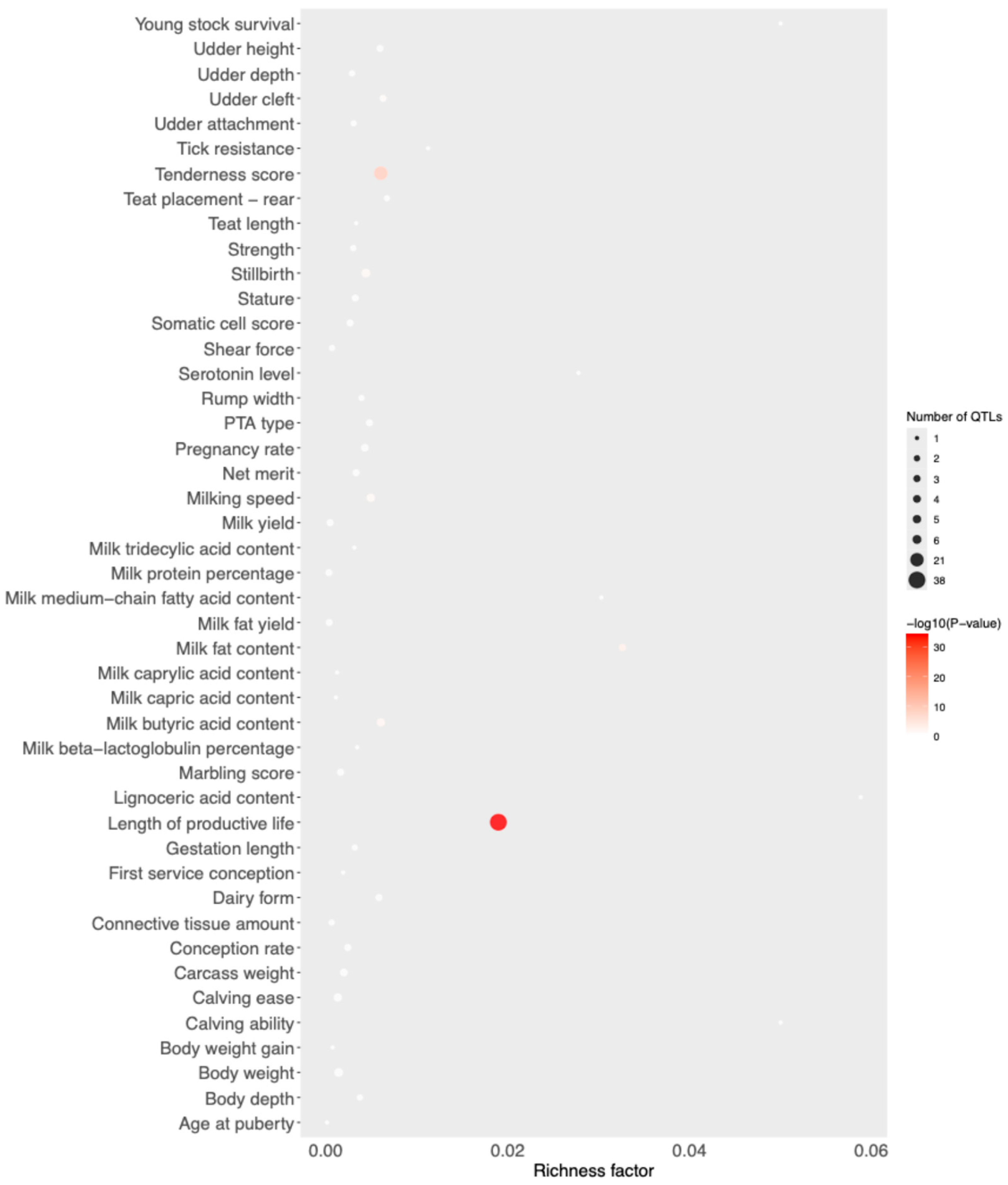
| Group | Breed | Code | Animals |
|---|---|---|---|
| Meat | Angus | ANG | 62 |
| Meat | Charolais | CHA | 46 |
| Meat | Hereford | HFD | 31 |
| Meat | Limousin | LMS | 44 |
| Meat | Piedmontese | PMT | 24 |
| Milk | Brown Swiss | BSW | 42 |
| Milk | Holstein | HOL | 63 |
| Milk | Jersey | JER | 49 |
| Milk | Montbéliarde | MON | 30 |
| Chromosome | Average FST Value | SNP FST | Average CDA Value | SNP CDA | Common SNPs |
|---|---|---|---|---|---|
| 1 | 0.079 ± 0.008 | 38 | −1.67 ± 8.21 | 12 | – |
| 2 | 0.076 ± 0.003 | 24 | 0.72 ± 3.72 | 6 | – |
| 3 | 0.086 ± 0.006 | 19 | −1.76 ± 3.78 | 10 | – |
| 4 | 0.084 ± 0.009 | 29 | −0.13 ± 9.39 | 16 | – |
| 5 | 0.109 ± 0.011 | 31 | 0.91 ± 8.36 | 7 | – |
| 6 | 0.078 ± 0.005 | 30 | 1.40 ± 7.73 | 18 | – |
| 7 | 0.086 ± 0.008 | 24 | −1.58 ± 6.92 | 14 | 1 |
| 8 | 0.073 ± 0.001 | 9 | 1.26 ± 7.63 | 12 | – |
| 9 | 0.076 ± 0.005 | 14 | −0.38 ± 1.7 | 9 | – |
| 10 | 0.095 ± 0.018 | 26 | 0.76 ± 6.17 | 10 | 2 |
| 11 | 0.095 ± 0.014 | 23 | 2.09 ± 3.19 | 8 | – |
| 12 | 0.070 ± 0.002 | 6 | −1.44 ± 5.36 | 8 | – |
| 13 | 0.114 ± 0.020 | 16 | 1.49 ± 8.99 | 17 | – |
| 14 | 0.120 ± 0.019 | 28 | 2.41 ± 5.98 | 9 | 1 |
| 15 | 0.077 ± 0.007 | 11 | −0.31 ± 2.72 | 11 | – |
| 16 | 0.088 ± 0.008 | 17 | 3.19 ± 10.58 | 15 | 2 |
| 17 | 0.079 ± 0.006 | 10 | 2.04 ± 5.74 | 10 | 1 |
| 18 | 0.087 ± 0.006 | 19 | 4.47 ± 4.09 | 7 | 2 |
| 19 | 0.093 ± 0.020 | 11 | 1.60 ± 3.86 | 9 | – |
| 20 | 0.089 ± 0.023 | 14 | −1.27 ± 7.41 | 12 | – |
| 21 | 0.138 ± 0.053 | 11 | −0.04 ± 5.22 | 8 | 1 |
| 22 | 0.071 ± 0.003 | 6 | 0.72 ± 5.79 | 11 | – |
| 23 | 0.095 ± 0.037 | 6 | −1.21 ± 4.04 | 15 | – |
| 24 | 0.084 ± 0.014 | 15 | 1.79 ± 5.50 | 8 | 1 |
| 25 | 0.060 ± 0.003 | 12 | 3.98 ± 4.97 | 7 | – |
| 26 | 0.087 ± 0.007 | 19 | −1.52 ± 6.00 | 6 | 1 |
| 27 | 0.088 ± 0.011 | 9 | 1.48 ± 6.13 | 8 | – |
| 28 | 0.086 ± 0.016 | 15 | 5.68 ± 9.83 | 6 | – |
| 29 | 0.079 ± 0.009 | 10 | −2.78 ± 6.93 | 6 | – |
| Total | 502 | 295 | 12 |
| BTA | SNP Name | Position | CDA Score | FST Smoothed Value |
|---|---|---|---|---|
| 7 | Hapmap53962-rs29017056 | 107,797,993 | 6.7711 | 0.0788 |
| 10 | ARS-BFGL-NGS-112081 | 36,489,310 | −1.1832 | 0.0750 |
| 10 | ARS-BFGL-NGS-34863 | 40,333,013 | −9.9492 | 0.0983 |
| 14 | ARS-BFGL-NGS-110022 | 38,481,264 | 7.8295 | 0.1158 |
| 16 | BTB-00639530 | 38,137,107 | 5.6524 | 0.0853 |
| 16 | ARS-BFGL-NGS-114895 | 38,205,760 | 33.8328 | 0.0940 |
| 17 | Hapmap44543-BTA-40914 | 39,220,916 | 14.5565 | 0.0821 |
| 18 | Hapmap47624-BTA-44484 | 12,578,668 | 0.9083 | 0.0931 |
| 18 | ARS-BFGL-NGS-100080 | 57,529,674 | 8.6108 | 0.0939 |
| 24 | BTB-00886858 | 34,750,786 | 7.6451 | 0.0959 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Congiu, M.; Cesarani, A.; Falchi, L.; Macciotta, N.P.P.; Dimauro, C. Combined Use of Univariate and Multivariate Approaches to Detect Selection Signatures Associated with Milk or Meat Production in Cattle. Genes 2024, 15, 1516. https://doi.org/10.3390/genes15121516
Congiu M, Cesarani A, Falchi L, Macciotta NPP, Dimauro C. Combined Use of Univariate and Multivariate Approaches to Detect Selection Signatures Associated with Milk or Meat Production in Cattle. Genes. 2024; 15(12):1516. https://doi.org/10.3390/genes15121516
Chicago/Turabian StyleCongiu, Michele, Alberto Cesarani, Laura Falchi, Nicolò Pietro Paolo Macciotta, and Corrado Dimauro. 2024. "Combined Use of Univariate and Multivariate Approaches to Detect Selection Signatures Associated with Milk or Meat Production in Cattle" Genes 15, no. 12: 1516. https://doi.org/10.3390/genes15121516
APA StyleCongiu, M., Cesarani, A., Falchi, L., Macciotta, N. P. P., & Dimauro, C. (2024). Combined Use of Univariate and Multivariate Approaches to Detect Selection Signatures Associated with Milk or Meat Production in Cattle. Genes, 15(12), 1516. https://doi.org/10.3390/genes15121516







