HOXA7 Expression Is an Independent Prognostic Biomarker in Esophageal Squamous Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of HOX Gene Expression Using Publicly Available Esophageal
2.2. Evaluation of Somatic Alterations in HOX Genes in the TCGA Data
2.3. Snap-Frozen Human Tissue Samples
2.4. Evaluation of Gene Expression by Quantitative PCR (qPCR)
2.5. Identification of HOX Targets and Gene-Set Enrichment Analyses
2.6. Statistical Analyses
3. Results
3.1. Mutational Profile of HOX Genes
3.2. HOX Genes’ Expression Profile
3.3. Targets of ESCC-Overexpressed HOX Genes Are Associated with Enriched Signaling Pathways and Cellular Processes
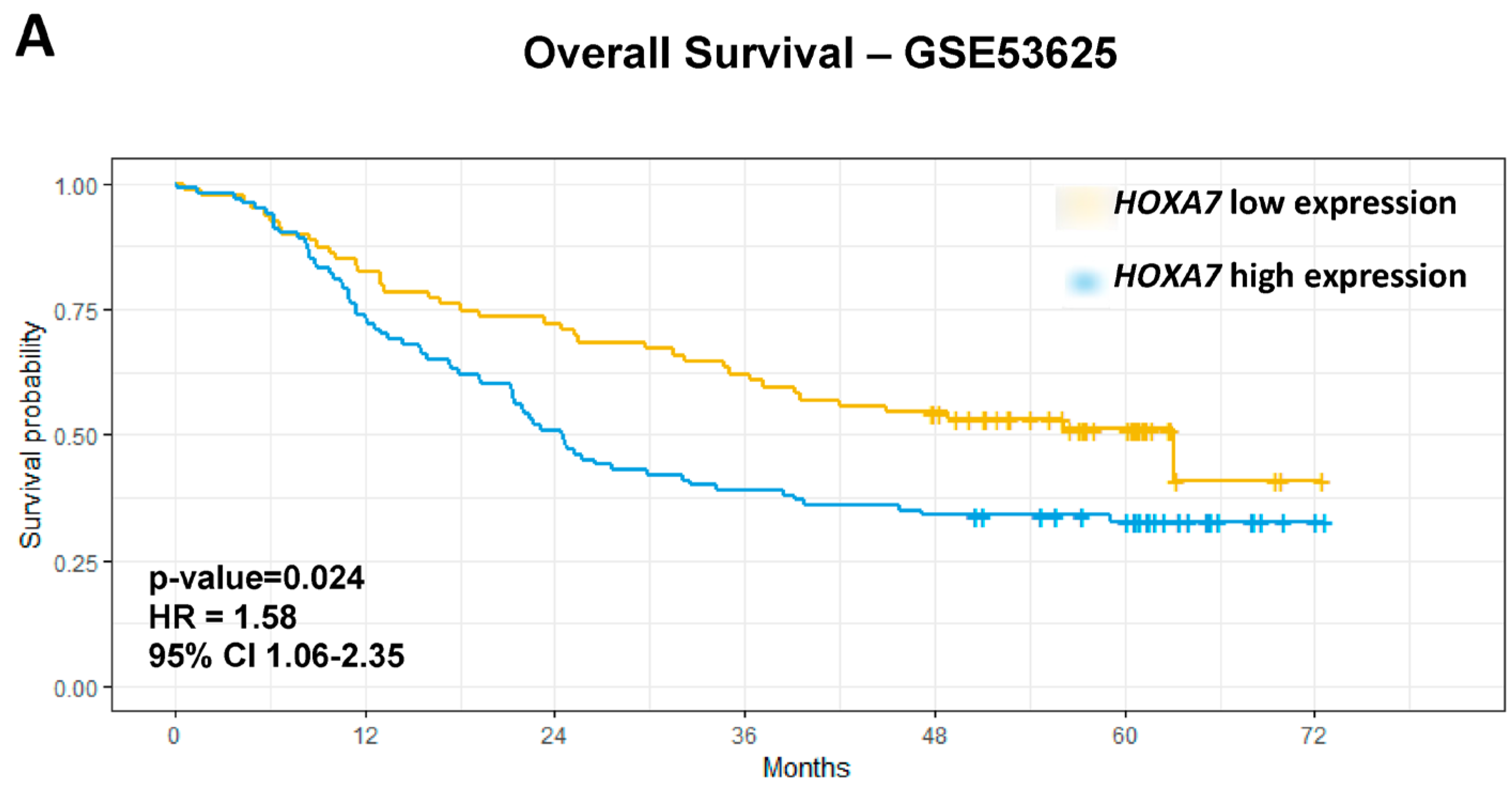
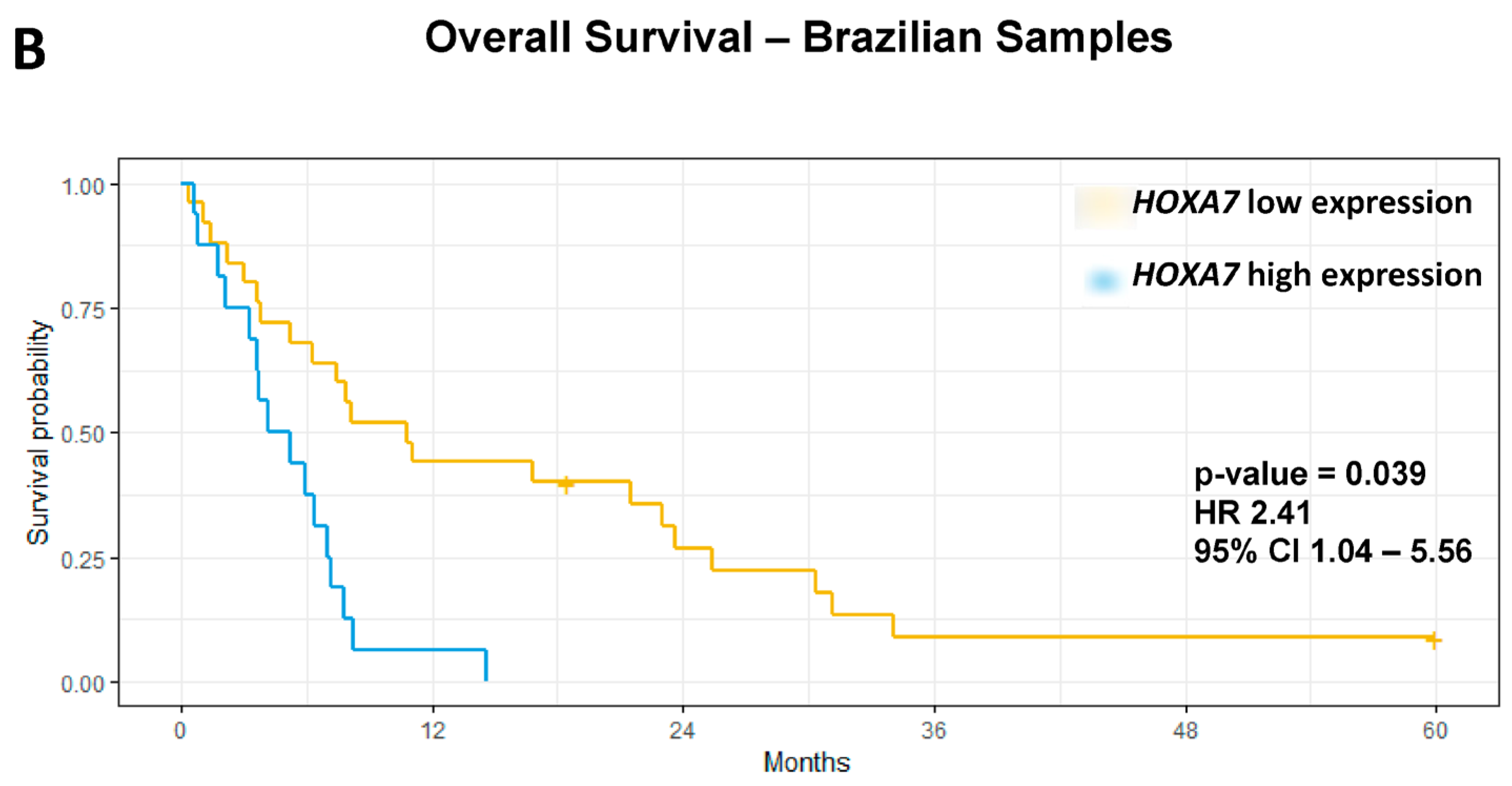
3.4. Association Analyses Between ESCC-Overexpressed HOX Genes and Clinicopathological Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewis, E.B. Clusters of master control genes regulate the development of higher organisms. JAMA 1992, 267, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sukumar, S. The HOX genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Krumlauf, R. HOX genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Kim, R.S.; Zhang Lee, S.J.; Jeng, M.H. HOXB13 induces growth suppression of prostate cancer cells as a hormone-activated androgen receptor signaling repressor. Cancer Res. 2004, 64, 9185–9191. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Dahiya, S.; Provencher, H.; Muir, B.; Carney, E.; Coser, K.; Shioda, T.; Ma, X.J.; Sgroi, D.C. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin. Cancer Res. 2007, 13, 6327–6334. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, B.; Mao, F.; Xu, J.; Miao, H.; Zou, Z.; Jang, Y.; Cai, S.; Witkin, M.; Koche, R.; et al. HOXA9 reprograms the enhancer landscape to promote leukemogenesis. Cancer Cell 2018, 34, 643–658. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. 2014, 92, 811–823. [Google Scholar] [CrossRef]
- Ewing, C.M.; Ray, A.M.; Lange, E.M.; Zuhlke, K.A.; Robbins, C.M.; Tembe, W.D.; Wiley, K.E.; Isaacs, S.D.; Johng, D.; Wang, Y.; et al. Germline mutations in HOXB13 and prostate-cancer risk. N. Engl. J. Med. 2012, 366, 141–149. [Google Scholar] [CrossRef]
- Luo, Z.; Rhie, S.K.; Farnham, P.J. The enigmatic HOX genes: Can we crack their code? Cancers 2019, 11, 323. [Google Scholar] [CrossRef]
- Chen, K.N.; Gu, Z.D.; Ke, Y.; Li, J.Y.; Shi, X.T.; Xu, G.W. Expression of 11 HOX genes is deregulated in esophageal squamous cell carcinoma. Clin. Cancer Res. 2005, 11, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Feng, A.; He, L.; Jiang, J.; Chu, Y.; Zhang, Z.; Fang, K.; Wang, Z.; Li, Z.; Sun, M.; Zhao, Z.; et al. Homeobox A7 promotes esophageal squamous cell carcinoma progression through C-C motif chemokine ligand 2-mediated tumor-associated macrophage recruitment. Cancer Sci. 2023, 114, 3270–3286. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, O.; Hamada, J.I.; Abe, M.; Hata, S.; Asano, T.; Takahashi, Y.; Tada, M.; Miyamoto, M.; Kondo, S.; Moriuchi, T. Dysregulated expression of HOX and ParaHOX genes in human esophageal squamous cell carcinoma. Oncol. Rep. 2007, 17, 753–760. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, J.; Jia, X.; Li, Q.; Zhang, H.; Wang, J.; Huang, S.; Hu, Z.; Li, C. Genomic and transcriptional characterization of early esophageal squamous cell carcinoma. BMC Med. Genom. 2023, 16, 67. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. 2024. Available online: https://gco.iarc.who.int/today (accessed on 30 August 2024).
- van Hagen, P.; Hulshof, M.C.C.M.; van Lanschot, J.J.B.; Steyerberg, E.W.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; Richel, D.J.; Nieuwenhuijzen, G.A.P.; Hospers, G.A.P.; Bonenkamp, J.J.; et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N. Engl. J. Med. 2012, 366, 2074–2084. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- Nicolau-Neto, P.; Da Costa, N.M.; de Souza Santos, P.T.; Gonzaga, I.M.; Ferreira, M.A.; Guaraldi, S.; Moreira, M.A.; Seuánez, H.N.; Brewer, L.; Bergmann, A.; et al. Esophageal squamous cell carcinoma transcriptome reveals the effect of FOXM1 on patient outcome through novel PIK3R3 mediated activation of PI3K signaling pathway. Oncotarget 2018, 9, 16634–16647. [Google Scholar] [CrossRef]
- Li, J.; Chen, Z.; Tian, L.; Zhou, C.; He, M.Y.; Gao, Y.; Wang, S.; Zhou, F.; Shi, S.; Feng, X.; et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 2014, 63, 1700–1710. [Google Scholar] [CrossRef]
- Sean, D.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar]
- Wettenhall, J.M.; Smyth, G.K. limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics 2004, 20, 3705–3706. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Plaisier, C.L.; O’Brien, S.; Bernard, B.; Reynolds, S.; Simon, Z.; Toledo, C.M.; Ding, Y.; Reiss, D.J.; Paddison, P.J.; Baliga, N.S. Causal mechanistic regulatory network for glioblastoma deciphered using systems genetics network analysis. Cell Syst. 2016, 3, 172–186. [Google Scholar] [CrossRef] [PubMed]
- John MYuxing, L.; Zhiao, S.; Qian, Z.; Alexander, R.P.; Bing, Z. WebGestalt 2024: Faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 2024, gkae456. [Google Scholar]
- Bradburn, M.J.; Clark, T.G.; Love, S.B.; Altman, D.G. Survival analysis part III: Multivariate data analysis—Choosing a model and assessing its adequacy and fit. Br. J. Cancer 2003, 89, 605–611. [Google Scholar] [CrossRef]
- Therneau, T.; Lumley, T. R Package, Version 2.38; Package Survival: A Package for Survival Analysis in R, 2015; Available online: https://cran.r-project.org/web/packages/survival/index.html (accessed on 1 October 2024).
- Lánczky, A.; Győrffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Hammerman, P.S.; Lawrence, M.S.; Voet, D.; Jing, R.; Cibulskis, K.; Sivachenko, A.; Stojanov, P.; McKenna, A.; Lander, E.S.; Gabriel, S.; et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef]
- Oguma, J.; Ozawa, S.; Kazuno, A.; Nitta, M.; Ninomiya, Y.; Kajiwara, H. Wnt3a expression is associated with poor prognosis of esophageal squamous cell carcinoma. Oncol. Lett. 2018, 15, 3100–3108. [Google Scholar] [CrossRef]
- de Souza-Santos, P.T.; Lima, S.C.S.; Nicolau-Neto, P.; Boroni, M.; Costa, N.M.; Brewer, L.; Menezes, A.N.; Furtado, C.; Moreira, M.A.M.; Seuanez, H.N.; et al. Mutations, differential gene expression, and chimeric transcripts in esophageal squamous cell carcinoma show high heterogeneity. Transl. Oncol. 2018, 11, 1283–1291. [Google Scholar] [CrossRef]
- Soares-Lima, S.C.; Gonzaga, I.M.; Camuzi, D.; Nicolau-Neto, P.; Silva, R.V.D.; Guaraldi, S.; Ferreira, M.A.; Hernandez-Vargas, H.; Herceg, Z.; Ribeiro Pinto, L.F. IL6 and BCL3 expression are potential biomarkers in esophageal squamous cell carcinoma. Front. Oncol. 2021, 11, 610574. [Google Scholar] [CrossRef] [PubMed]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Yu, J.; Zhao, S.; Cao, X.; Wang, Q. HOXA7 promotes the metastasis of KRAS mutant colorectal cancer by regulating myeloid-derived suppressor cells. Cancer Cell Int. 2022, 22, 89. [Google Scholar] [CrossRef] [PubMed]
- Padam, K.S.R.; Morgan, R.; Hunter, K.; Chakrabarty, S.; Kumar, N.A.N.; Radhakrishnan, R. Identification of HOX signatures contributing to oral cancer phenotype. Sci. Rep. 2022, 12, 2457. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.H.; Fang, S.J.; Qin, C.F.; Sun, R.L.; Liu, Z.Y.; Jiang, B.Y.; Wu, X.; Li, G. HOXA7 stimulates human hepatocellular carcinoma proliferation through cyclin E1/CDK2. Oncol. Rep. 2015, 33, 990–996. [Google Scholar] [CrossRef]
- Duan, X.; Chen, H.; Ma, H.; Song, Y. The expression and significance of the HOXA7 gene in oral squamous cell carcinoma. J. Oral Sci. 2017, 59, 329–335. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Q.; Cheng, J.C.; Nishi, Y.; Yanase, T.; Huang, H.F.; Leung, P.C. Homeobox A7 increases cell proliferation by up-regulation of epidermal growth factor receptor expression in human granulosa cells. Reprod. Biol. Endocrinol. 2010, 8, 61. [Google Scholar] [CrossRef]
- Hu, X.; Wang, Y.; Zhang, X.; Li, C.; Zhang, X.; Yang, D.; Liu, Y.; Li, L. DNA methylation of HOX genes and its clinical implications in cancer. Exp. Mol. Pathol. 2023, 134, 104916. [Google Scholar] [CrossRef]
- Sun, M.; Song, C.X.; Huang, H.; Frankenberger, C.A.; Sankarasharma, D.; Gomes, S.; Chen, P.; Chen, J.; Chada, K.K.; He, C.; et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9920–9925. [Google Scholar] [CrossRef]

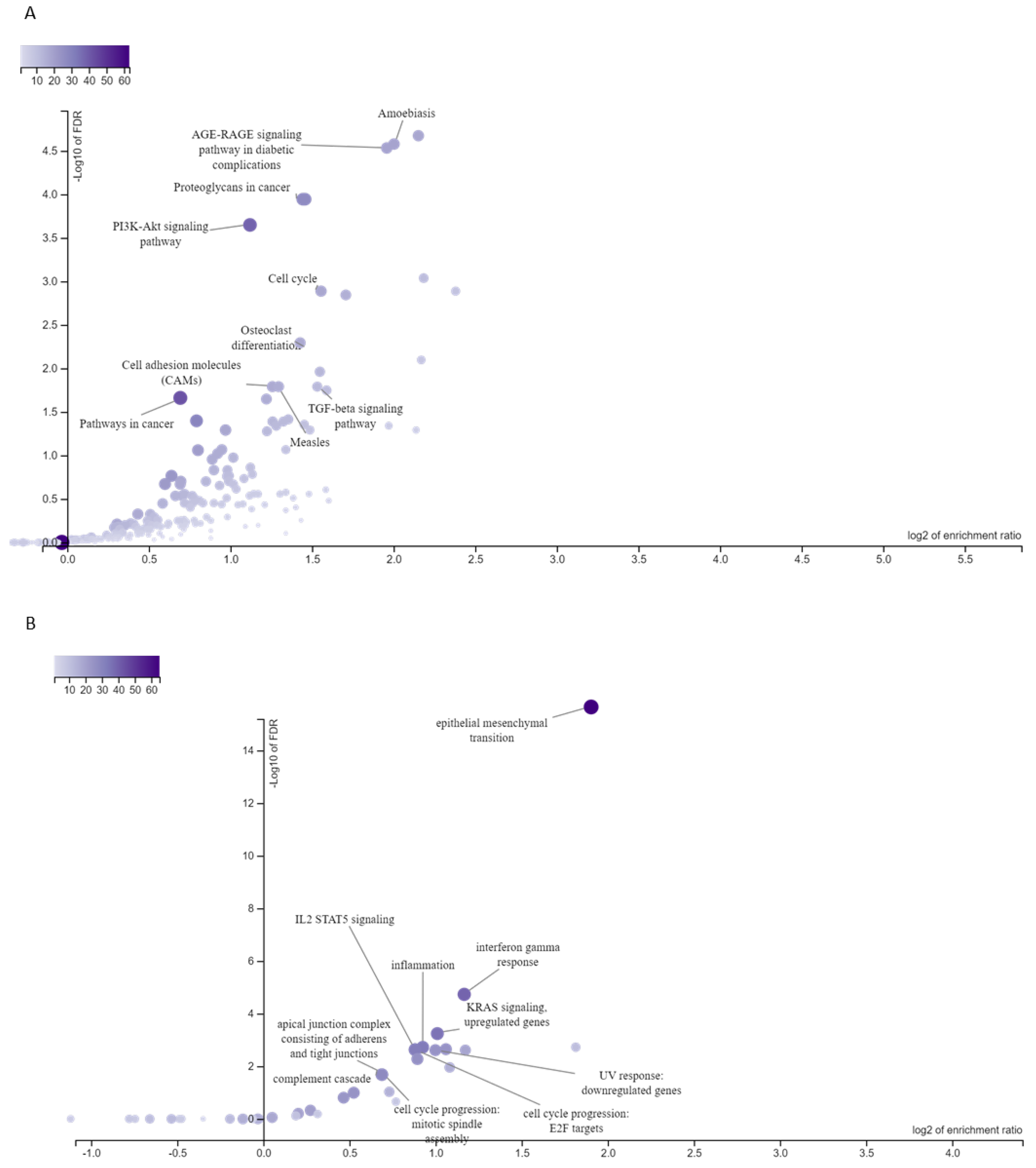
| Feature | Variable | Brazilian ESCC Patients |
|---|---|---|
| Age—Median (range) | 59 (39–77) | |
| Gender | Female | 8 (19.51%) |
| Male | 33 (80.49%) | |
| Tobacco smoking | No | 4 (9.76%) |
| Yes | 35 (85.36%) | |
| NA | 2 (4.88%) | |
| Alcohol drinking | No | 5 (12.20%) |
| Yes | 35 (85.36%) | |
| NA | 1 (2.44%) | |
| Esophagel tumor subsite | Upper | 5 (12.20%) |
| Middle | 11 (26.83%) | |
| Lower | 3 (7.3%) | |
| More than one subsite | 22 (53.66%) | |
| Tumor grade | G2 | 34 (82.93%) |
| G3 | 7 (17.07%) | |
| Stage | I-II | 12 (29.27%) |
| III-IV | 25 (60.97%) | |
| NA | 4 (9.76%) | |
| Lymph node metastasis | No | 8 (19.51%) |
| Yes | 17 (41.46%) | |
| NA | 16 (39.03%) | |
| Distant metastasis | No | 11 (26.83%) |
| Yes | 17 (41.46%) | |
| NA | 13 (31.71%) |
| Gene Symbol | GSE75241 | GSE53625 | ||||
|---|---|---|---|---|---|---|
| Probe ID | Fold Change (ESCC/NMSM) | p Value | Probe ID | Fold Change (ESCC/NMSM) | p Value | |
| HOXA2 | 3042756 | −1.67 | <0.001 | CB_015587 | −1.58 | <0.001 |
| HOXA7 | 3042881 | 1.50 | 0.002 | CB_015737 | 2.13 | <0.001 |
| HOXB13 | 3761538 | 2.12 | <0.001 | CB_015218 | 2.87 | <0.001 |
| HOXC9 | 3416344 | 1.57 | <0.001 | CB_015738 | 3.43 | <0.001 |
| HOXC10 | 3416290 | 2.32 | <0.001 | CB_019037 | 10.56 | <0.001 |
| HOXC13 | 3416256 | 1.66 | <0.001 | CB_019038 | 15.45 | <0.001 |
| HOXD10 | 2516834 | 4.17 | <0.001 | CB_011132 | 4.44 | <0.001 |
| Frequency | HOXA2 | HOXA7 | HOXB13 | HOXC9 | HOXC10 | HOXC13 | HOXD10 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Feature | Variable | n | % | Median | p Value | Median | p Value | Median | p Value | Median | p Value | Median | p Value | Median | p Value | Median | p Value |
| Age | <60 | 89 | 49.70% | 10.54 | 0.11 | 12.94 | 0.22 | 5.76 | 0.026 | 10.70 | 0.16 | 9.59 | 0.85 | 9.14 | 0.38 | 11.89 | 0.92 |
| ≥60 | 90 | 50.30% | 10.83 | 12.87 | 6.83 | 10.91 | 9.64 | 9.18 | 11.88 | ||||||||
| Sex | Male | 146 | 81.60% | 10.78 | 0.24 | 12.95 | 0.12 | 6.53 | 0.78 | 10.83 | 0.50 | 10.14 | 0.88 | 9.13 | 0.67 | 11.97 | 0.009 |
| Female | 33 | 18.40% | 10.61 | 12.73 | 6.15 | 10.7 | 9.95 | 9.24 | 11.50 | ||||||||
| Alcohol Drinking | No | 73 | 40.80% | 10.77 | 0.65 | 12.74 | 0.09 | 6.24 | 0.73 | 10.65 | 0.09 | 9.94 | 0.66 | 8.98 | 0.06 | 11.80 | 0.01 |
| Yes | 106 | 59.20% | 10.73 | 13.04 | 6.32 | 10.91 | 10.22 | 9.43 | 12.11 | ||||||||
| Tobacco Smoking | No | 65 | 36.30% | 10.73 | 0.8 | 12.82 | 0.33 | 5.84 | 0.41 | 10.53 | 0.005 | 9.99 | 0.92 | 9.05 | 0.27 | 11.88 | 0.53 |
| Yes | 114 | 63.70% | 10.76 | 12.97 | 6.82 | 10.97 | 10.16 | 9.27 | 12.00 | ||||||||
| Esophageal Tumor Subsite | Upper | 20 | 11.20% | 10.96 | 0.81 | 13.34 | 0.09 | 6.87 | 0.62 | 11.23 | 0.36 | 9.88 | 0.70 | 8.88 | 0.3 | 11.76 | 0.48 |
| Middle | 97 | 54.20% | 10.62 | 12.98 | 6.69 | 10.96 | 10.00 | 9.09 | 12.06 | ||||||||
| Lower | 62 | 34.60% | 10.62 | 12.66 | 6.87 | 10.55 | 10.39 | 9.43 | 11.81 | ||||||||
| Tumor Differentiation | Well | 32 | 17.90% | 10.59 | 0.68 | 13.11 | 0.74 | 5.66 | 0.0501 | 10.56 | 0.0067 * | 9.89 | 0.47 | 9.48 | 0.92 | 11.8 | 0.75 |
| Moderate | 98 | 54.70% | 10.61 | 12.89 | 6.32 | 10.72 | 9.94 | 9.15 | 12.03 | ||||||||
| Poorly | 49 | 27.40% | 10.79 | 13.03 | 7.36 | 11.27 | 10.72 | 9.08 | 12.05 | ||||||||
| Lymph node metastasis | No | 83 | 46.40% | 10.68 | 0.55 | 12.83 | 0.74 | 6.87 | 0.31 | 10.80 | 0.89 | 10.00 | 0.72 | 9.06 | 0.30 | 11.95 | 0.95 |
| Yes | 96 | 53.60% | 10.62 | 12.95 | 5.73 | 10.82 | 10.25 | 9.40 | 11.97 | ||||||||
| T (TNM) | T1/T2 | 39 | 21.70% | 10.54 | 0.3 | 12.83 | 0.82 | 6.35 | 0.65 | 11.01 | 0.16 | 10.34 | 0.65 | 9.40 | 0.39 | 11.91 | 0.84 |
| T3/T4 | 140 | 78.80% | 10.66 | 12.94 | 6.27 | 10.75 | 10.00 | 9.10 | 11.88 | ||||||||
| Tumor Stage | Early (I/II) | 87 | 48.60% | 10.57 | 0.08 | 12.82 | 0.55 | 6.87 | 0.09 | 10.77 | 0.61 | 9.63 | 0.95 | 9.08 | 0.37 | 11.88 | 0.95 |
| Late (III) | 92 | 51.40% | 10.68 | 13.01 | 5.65 | 10.84 | 9.61 | 9.27 | 12.05 | ||||||||
| Feature | Variable | GSE53625 | Brazilian Samples | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Survival Analysis | Multivariate Analysis | Univariate Survival Analysis | Multivariate Analysis | ||||||||||||||
| 95% CI | 95% CI | 95% CI | 95% CI | ||||||||||||||
| HR | Low | High | p Value | HR | Low | High | p Value | HR | Low | High | p Value | HR | Low | High | p Value | ||
| Age | ≥60 y vs. <60 y | 1.54 | 1.047 | 2.26 | 0.028 | 0.98 | 0.51 | 1.89 | 0.97 | ||||||||
| Gender | Male vs. Female | 0.78 | 0.49 | 1.25 | 0.3 | 2.97 | 1.4 | 3.12 | 0.0002 | 0.64 | 0.27 | 1.51 | 0.31 | ||||
| Stage | I vs. II vs. III | 2.15 | 1.44 | 3.2 | 0.00015 | 3.02 | 1.27 | 7.19 | 0.012 | 2.36 | 0.93 | 5.96 | 0.06 | ||||
| Grade | G3/G2 vs. G1 | 1.35 | 1 | 1.82 | 0.048 | 0.57 | 0.28 | 1.17 | 0.12 | ||||||||
| HOXA2 | High vs. Low | 1.58 | 1.041 | 2.408 | 0.0316 | ||||||||||||
| HOXA7 | High vs. Low | 1.65 | 1.11 | 2.45 | 0.013 | 1.58 | 1.06 | 2.35 | 0.024 | 3.29 | 1.54 | 6.99 | 0.0019 | 2.41 | 1.04 | 5.56 | 0.039 |
| HOXB13 | High vs. Low | 1.9 | 1.06 | 3.4 | 0.0302 | ||||||||||||
| HOXC9 | High vs. Low | 1.52 | 0.96 | 2.42 | 0.073 | ||||||||||||
| HOXC10 | High vs. Low | 0.28 | 0.14 | 1.04 | 0.061 | ||||||||||||
| HOXC13 | High vs. Low | 1.28 | 0.87 | 1.88 | 0.13 | ||||||||||||
| HOXD10 | High vs. Low | 1.3 | 0.87 | 1.9 | 0.018 | ||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, J.V.; Nicolau-Neto, P.; de Almeida, J.N.; Lisboa, L.B.; de Souza-Santos, P.T.; Ribeiro-Pinto, L.F.; Soares-Lima, S.C.; Simão, T.d.A. HOXA7 Expression Is an Independent Prognostic Biomarker in Esophageal Squamous Cell Carcinoma. Genes 2024, 15, 1430. https://doi.org/10.3390/genes15111430
Gomes JV, Nicolau-Neto P, de Almeida JN, Lisboa LB, de Souza-Santos PT, Ribeiro-Pinto LF, Soares-Lima SC, Simão TdA. HOXA7 Expression Is an Independent Prognostic Biomarker in Esophageal Squamous Cell Carcinoma. Genes. 2024; 15(11):1430. https://doi.org/10.3390/genes15111430
Chicago/Turabian StyleGomes, Jennifer Vieira, Pedro Nicolau-Neto, Júlia Nascimento de Almeida, Lilian Brewer Lisboa, Paulo Thiago de Souza-Santos, Luis Felipe Ribeiro-Pinto, Sheila Coelho Soares-Lima, and Tatiana de Almeida Simão. 2024. "HOXA7 Expression Is an Independent Prognostic Biomarker in Esophageal Squamous Cell Carcinoma" Genes 15, no. 11: 1430. https://doi.org/10.3390/genes15111430
APA StyleGomes, J. V., Nicolau-Neto, P., de Almeida, J. N., Lisboa, L. B., de Souza-Santos, P. T., Ribeiro-Pinto, L. F., Soares-Lima, S. C., & Simão, T. d. A. (2024). HOXA7 Expression Is an Independent Prognostic Biomarker in Esophageal Squamous Cell Carcinoma. Genes, 15(11), 1430. https://doi.org/10.3390/genes15111430






