A PCR-Based Approach for Early Diagnosis of Head and Neck Aspergillosis: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
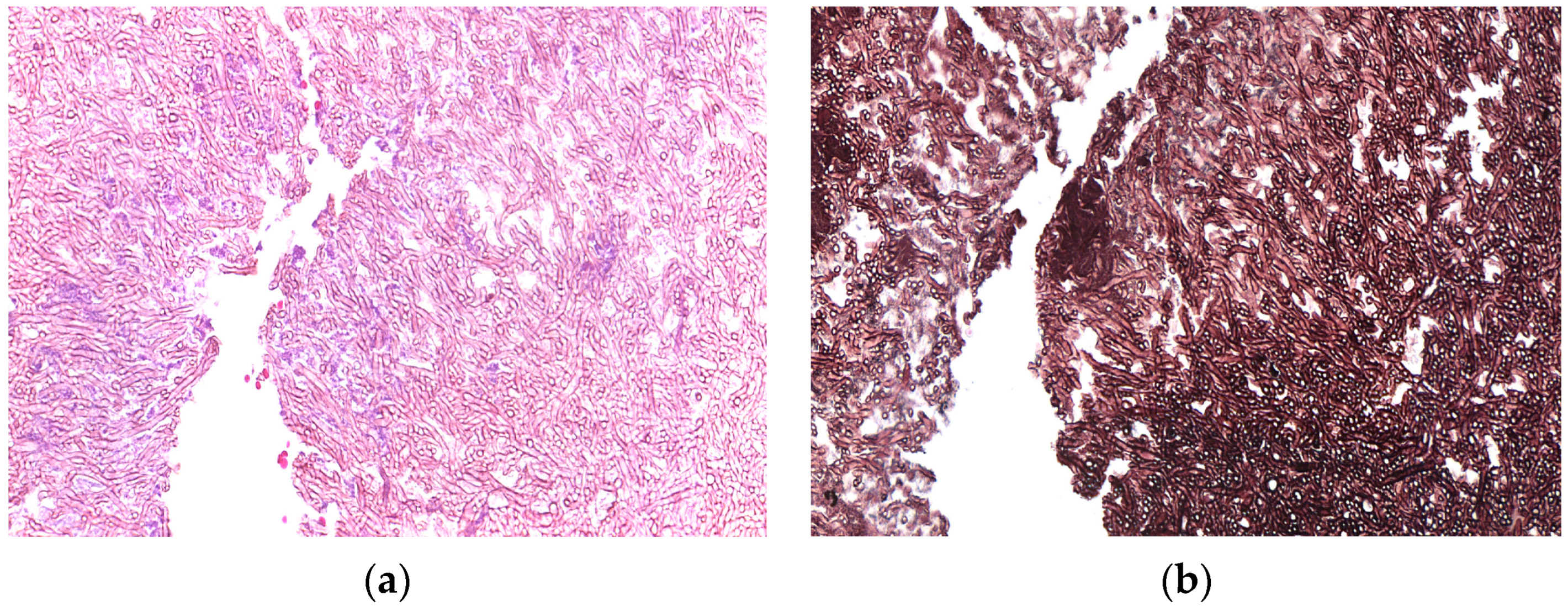
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker, S.E.; Bennett, J.W. An overview of the genus aspergillus. In The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods; CRC Press: Boca Raton, FL, USA, 2007; pp. 3–13. [Google Scholar]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2016, 10, 296–322. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5328810/ (accessed on 5 December 2023). [CrossRef] [PubMed]
- Batista, P.P. Caracterização de Linhagens do grupo Aspergillus flavus Baseada em Marcadores de DNA. repositorio.ufpe.br. 2007. Available online: https://repositorio.ufpe.br/handle/123456789/6454 (accessed on 5 December 2023).
- Egger, M.; Jenks, J.D.; Hoenigl, M.; Prattes, J. Blood Aspergillus PCR: The Good, the Bad, and the Ugly. J. Fungi 2020, 6, 18. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7151127/ (accessed on 17 February 2024). [CrossRef] [PubMed]
- Telles, D.R.; Karki, N.; Marshall, M.W. Oral Fungal Infections: Diagnosis and Management. Dent. Clin. N. Am. 2017, 61, 319–349. Available online: https://www.sciencedirect.com/science/article/abs/pii/S0011853216301380 (accessed on 22 February 2024). [CrossRef]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. Available online: https://cmr.asm.org/content/cmr/12/2/310.full.pdf (accessed on 11 March 2024). [CrossRef] [PubMed]
- Knoke, M.; Bernhardt, H.; Schwesinger, G. Early description of a pulmonary aspergillosis 1847 from Greifswald. Mycoses 2003, 46 (Suppl. S1), 37–41. Available online: https://pubmed.ncbi.nlm.nih.gov/12955852/ (accessed on 11 March 2024). [CrossRef]
- Cadena, J.; Thompson, G.R.; Patterson, T.F. Aspergillosis. Infect. Dis. Clin. N. Am. 2021, 35, 415–434. [Google Scholar] [CrossRef]
- Bongomin, F.; Harris, C.; Foden, P.; Kosmidis, C.; Denning, D.W. Innate and Adaptive Immune Defects in Chronic Pulmonary Aspergillosis. J. Fungi. 2017, 3, 26. [Google Scholar] [CrossRef]
- Sasani, E.; Pakdel, F.; Khodavaisy, S.; Salehi, M.; Salami, A.; Sohrabi, M.; Aminishakiba, P.; Amirafzali, I.; Khaneshan, A.S. Mixed Aspergillosis and Mucormycosis Infections in Patients with COVID-19: Case Series and Literature Review. Mycopathologia 2024, 189, 10. [Google Scholar] [CrossRef]
- Carvalho-Pereira, J.; Fernandes, F.; Araújo, R.; Springer, J.; Loeffler, J.; Buitrago, M.J.; Pais, C.; Sampaio, P. Multiplex PCR Based Strategy for Detection of Fungal Pathogen DNA in Patients with Suspected Invasive Fungal Infections. J. Fungi 2020, 6, 308. [Google Scholar] [CrossRef]
- Lortholary, O.; Gangneux, J.P.; Sitbon, K.; Lebeau, B.; de Monbrison, F.; Le Strat, Y.; Coignard, B.; Dromer, F.; Bretagne, S.; for the French Mycosis Study Group. Epidemiological trends in invasive aspergillosis in France: The SAIF network (2005–2007). Clin. Microbiol. Infect. 2011, 17, 1882–1889. [Google Scholar] [CrossRef]
- Migott, G.B.; dos Santos, F.M.; Pagnussat, L.R.; Barbosa, B.; Barbosa, G.D.L.; Hahn, S.R. Perfil Clínico e Epidemiológico de Pacientes com Suspeita de Aspergilose Pulmonar em Hospital do Estado Rio Grande do Sul, Brasil. Rev. Epidemiol. E Controle Infecção 2017, 7, 34–39. [Google Scholar] [CrossRef][Green Version]
- Rai, D.; Shukla, D.; Bhola, N.D. Aspergillosis of Maxillary Sinus’s Diagnosis, Management, and Association with COVID-19: A Case Report. Cureus 2022, 14, e30191. [Google Scholar] [CrossRef]
- Gomes, C.C.; Pinto, L.C.C.; Victor, F.L.; da Silva, E.A.B.; Ribeiro, A.d.A.; Sarquis, M.I.d.M.; Camões, I.C.G. Aspergillus in endodontic infection near the maxillary sinus. Braz. J. Otorhinolaryngol. 2015, 81, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; Pasini, R.A.; Pasqualotto, A.; Marchiori, E.; Altmayer, S.; Irion, K.; Mançano, A.; Hochhegger, B. Imaging Findings in Aspergillosis: From Head to Toe. Mycopathologia 2023, 188, 623–641. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.J.; Anaissie, E.J.; Denning, D.W.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Segal, B.H.; Steinbach, W.J.; Stevens, D.A.; et al. Treatment of aspergillosis: Clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 327–360. Available online: https://pubmed.ncbi.nlm.nih.gov/18177225/ (accessed on 5 June 2024). [CrossRef] [PubMed]
- Caggiano, G.; Apollonio, F.; Consiglio, M.; Gasparre, V.; Trerotoli, P.; Diella, G.; Lopuzzo, M.; Triggiano, F.; Stolfa, S.; Mosca, A.; et al. Tendency in Pulmonary Aspergillosis Investigation during the COVID-19 Era: What Is Changing? Int. J. Environ. Res. Public Health 2022, 19, 7079. [Google Scholar] [CrossRef]
- Balajee, S.A.; Marr, K.A. Phenotypic and Genotypic Identification of Human Pathogenic as Pergilli. Futur. Microbiol. 2006, 1, 435–445. [Google Scholar] [CrossRef]
- Steinbach, W.J. Are We There Yet? Recent Progress in the Molecular Diagnosis and Novel Antifungal Targeting of Aspergillus fumigatus and Invasive Aspergillosis. PLoS Pathog. 2013, 9, e1003642. [Google Scholar] [CrossRef]
- O’Gorman, C.M.; Fuller, H.T.; Dyer, P.S. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 2009, 457, 471–474. [Google Scholar] [CrossRef]
- Yang, S.; Rothman, R.E. PCR-based Diagnostics for Infectious diseases: Uses, limitations, and Future Applications in acute-care Settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Abate, M.S.; Battle, L.R.; Emerson, A.N.; Gardner, J.M.; Shalin, S.C. Dermatologic Urgencies and Emergencies: What Every Pathologist Should Know. Arch. Pathol. Lab. Med. 2019, 143, 919–942. [Google Scholar] [CrossRef] [PubMed]
- Ozhak-Baysan, B.; Alastruey-Izquierdo, A.; Saba, R.; Ogunc, D.; Ongut, G.; Timuragaoglu, A.; Arslan, G.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. Aspergillus alliaceus and Aspergillus flavus co-infection in an acute myeloid leukemia patient. Med. Mycol. 2010, 48, 995–999. Available online: https://academic.oup.com/mmy/article/48/7/995/1056456 (accessed on 14 March 2024). [CrossRef] [PubMed]


| Species | Primer Sequence (5′-3′) | Amplicon Size |
|---|---|---|
| A. fumigatus | F-GTCTGAGTTGATTATCGT | 312 bp |
| R-GGCCTACAGAGCAGGTGAC | ||
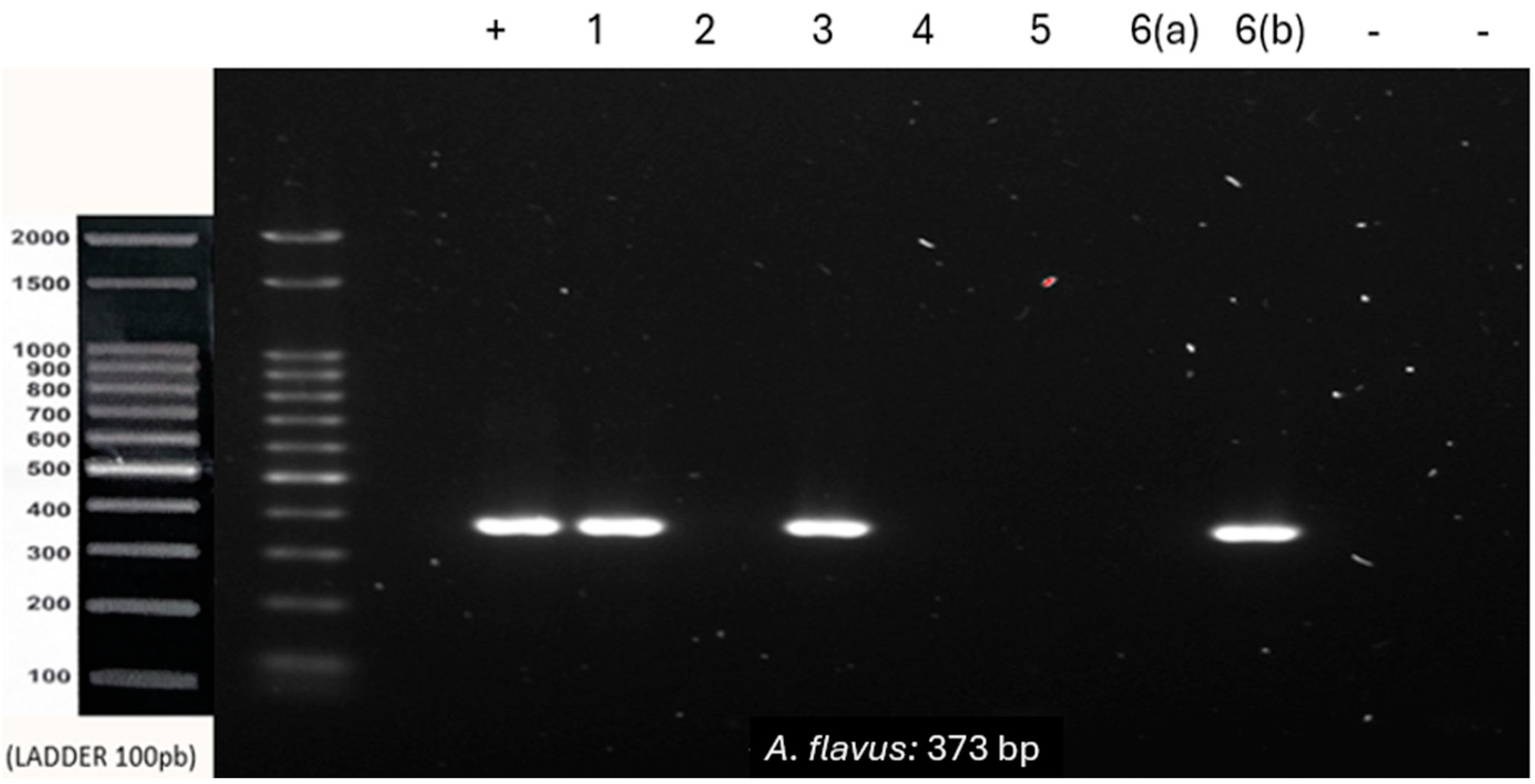
| A. flavus | F-CACCACGAACTCTGTCTGATC | 373 bp |
| R-GATTGATTTGCGTTCGGC | ||
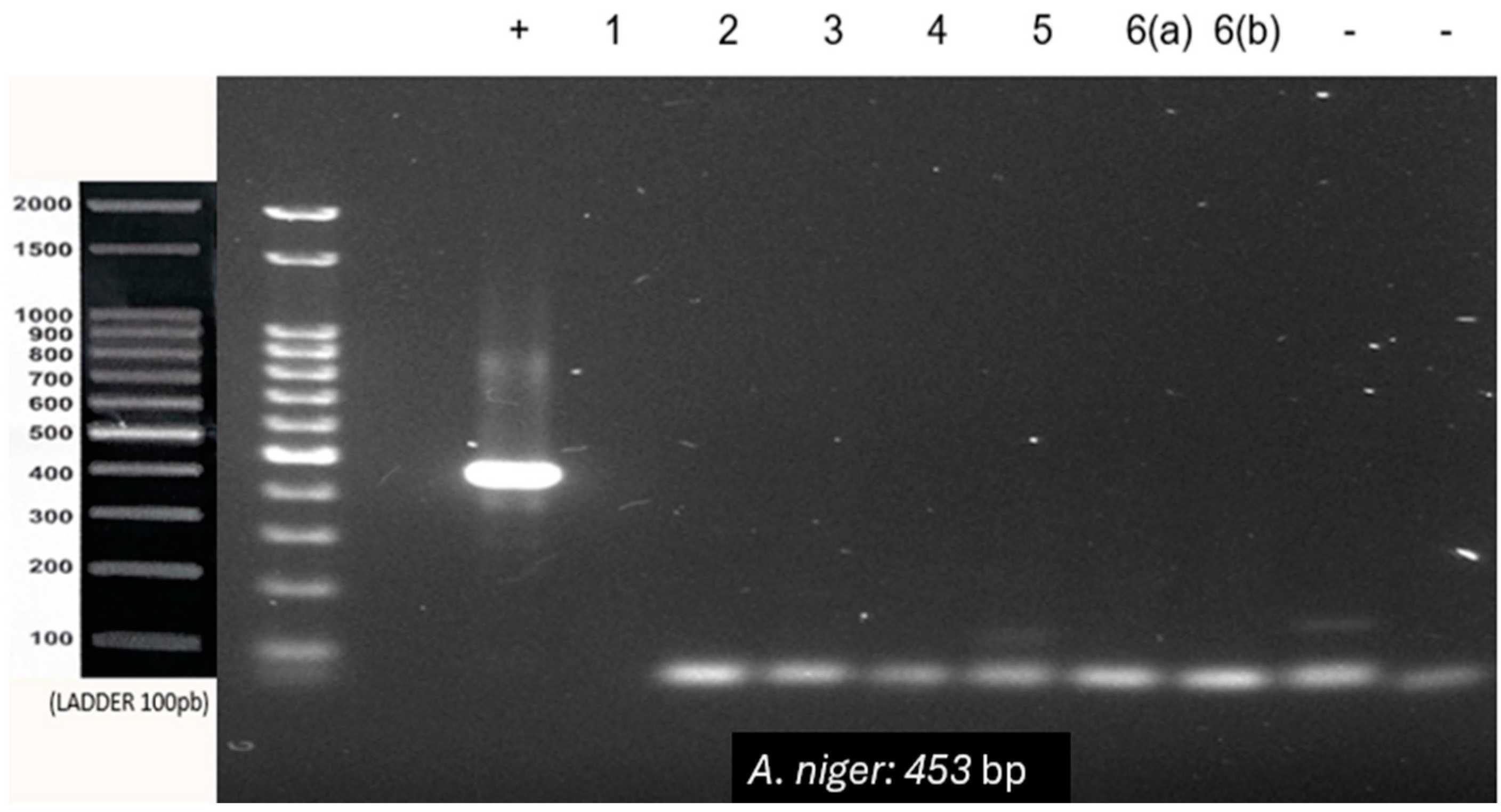
| A. niger | F-GCCCAACCTCCCATCCGTG | 453 bp |
| R-CAATCCTACAGAGCATGTG |
| Samples | Location | Sex | Age | PCR Test Results | ||
|---|---|---|---|---|---|---|
| A. niger | A. fumigatus | A. flavus | ||||
| 1 | Maxillary sinus | M | 79 | - | + | + |
| 2 | Maxillary sinus | M | 55 | - | - | - |
| 3 | Maxillary sinus | M | 49 | - | - | + |
| 4 | Upper gum | M | 56 | - | + | - |
| 5 | Frontal sinus | M | 57 | - | - | - |
| 6 | Maxillary sinus | M | 52 | - | - | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, T.E.C.; Bastos, V.C.; Boniek, D.; Romañach, M.; Rocha, F.F.; Chaves, R.R.M.; Gomez, R.S. A PCR-Based Approach for Early Diagnosis of Head and Neck Aspergillosis: A Pilot Study. Genes 2024, 15, 1428. https://doi.org/10.3390/genes15111428
Gomes TEC, Bastos VC, Boniek D, Romañach M, Rocha FF, Chaves RRM, Gomez RS. A PCR-Based Approach for Early Diagnosis of Head and Neck Aspergillosis: A Pilot Study. Genes. 2024; 15(11):1428. https://doi.org/10.3390/genes15111428
Chicago/Turabian StyleGomes, Thaís Ellen Chaves, Victor Coutinho Bastos, Douglas Boniek, Mário Romañach, Fernanda Faria Rocha, Roberta Rayra Martins Chaves, and Ricardo Santiago Gomez. 2024. "A PCR-Based Approach for Early Diagnosis of Head and Neck Aspergillosis: A Pilot Study" Genes 15, no. 11: 1428. https://doi.org/10.3390/genes15111428
APA StyleGomes, T. E. C., Bastos, V. C., Boniek, D., Romañach, M., Rocha, F. F., Chaves, R. R. M., & Gomez, R. S. (2024). A PCR-Based Approach for Early Diagnosis of Head and Neck Aspergillosis: A Pilot Study. Genes, 15(11), 1428. https://doi.org/10.3390/genes15111428








