LCM-RNAseq Highlights Intratumor Heterogeneity and a lncRNA Signature from Archival Tissues of GH-Secreting PitNETs
Abstract
1. Introduction
2. Methods
2.1. Patients’ Cohort
2.2. Sample Collection and Preparation
2.3. Preprocessing of RNA Sequencing Data
2.4. Quality Assessment
2.5. Data Transformation and Visualization
2.6. Gene Expression Analysis by Droplet Digital PCR (ddPCR)
3. Results
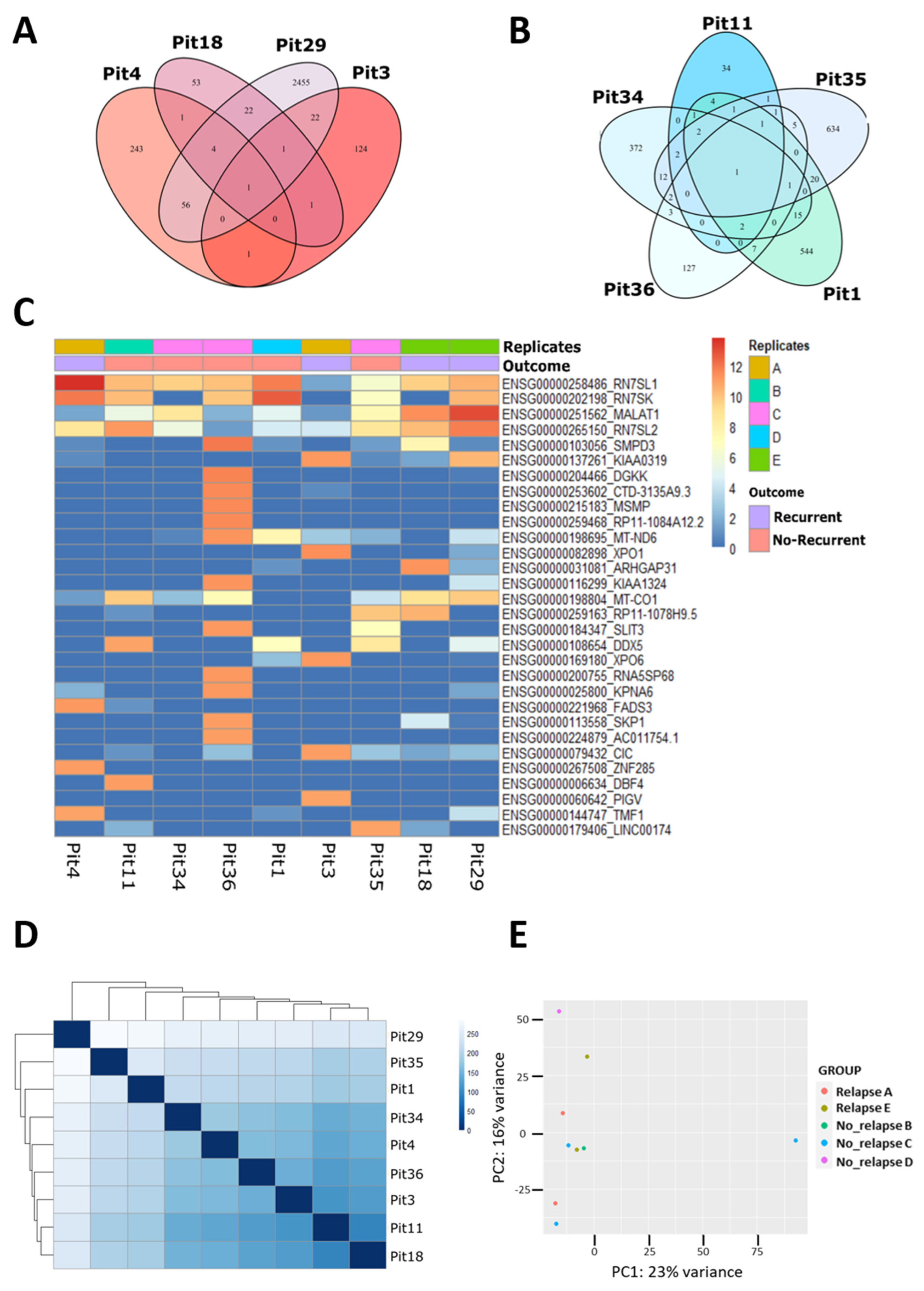
3.1. Transcriptomic Profile from FFPE GH-Secreting-PitNET by LCM-RNA-Seq
3.2. The lncRNAs in GH-Secreting PitNETs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lloyd, R.V.; Qian, X.; Jin, L.; Ruebel, K.; Bayliss, J.; Zhang, S.; Kobayashi, I. Analysis of Pituitary Cells by Laser Capture Microdissection. In Laser Capture Microdissection; Murray, G.I., Curran, S., Eds.; Humana Press: Totowa, NJ, USA, 2005; pp. 233–241. ISBN 978-1-58829-260-5. [Google Scholar]
- Nakamura, N.; Ruebel, K.; Jin, L.; Qian, X.; Zhang, H.; Lloyd, R.V. Laser Capture Microdissection for Analysis of Single Cells. In Single Cell Diagnostics; Thornhill, A., Ed.; Methods in Molecular MedicineTM; Humana Press: Totowa, NJ, USA, 2007; Volume 132, pp. 11–18. ISBN 978-1-58829-578-1. [Google Scholar]
- Neou, M.; Villa, C.; Armignacco, R.; Jouinot, A.; Raffin-Sanson, M.-L.; Septier, A.; Letourneur, F.; Diry, S.; Diedisheim, M.; Izac, B.; et al. Pangenomic Classification of Pituitary Neuroendocrine Tumors. Cancer Cell 2020, 37, 123–134.e5. [Google Scholar] [CrossRef] [PubMed]
- Inoshita, N.; Nishioka, H. The 2017 WHO Classification of Pituitary Adenoma: Overview and Comments. Brain Tumor. Pathol. 2018, 35, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Mete, O.; Lopes, M.B. Overview of the 2017 WHO Classification of Pituitary Tumors. Endocr. Pathol. 2017, 28, 228–243. [Google Scholar] [CrossRef]
- Marrero-Rodríguez, D.; Moscona-Nissan, A.; Sidauy-Adissi, J.; Haidenberg-David, F.; Jonguitud-Zumaya, E.; de Jesus Chávez-Vera, L.; Martinez-Mendoza, F.; Taniguchi-Ponciano, K.; Mercado, M. The Molecular Biology of Sporadic Acromegaly. Best Pract. Res. Clin. Endocrinol. Metab. 2024, 38, 101895. [Google Scholar] [CrossRef] [PubMed]
- Fleseriu, M.; Langlois, F.; Lim, D.S.T.; Varlamov, E.V.; Melmed, S. Acromegaly: Pathogenesis, Diagnosis, and Management. Lancet Diabetes Endocrinol. 2022, 10, 804–826. [Google Scholar] [CrossRef]
- Lenders, N.F.; McCormack, A.I.; Ho, K.K.Y. Management of Endocrine Disease: Does Gender Matter in the Management of Acromegaly? Eur. J. Endocrinol. 2020, 182, R67–R82. [Google Scholar] [CrossRef]
- Melmed, S. Acromegaly Pathogenesis and Treatment. J. Clin. Investig. 2009, 119, 3189–3202. [Google Scholar] [CrossRef]
- Stueven, A.K.; Kayser, A.; Wetz, C.; Amthauer, H.; Wree, A.; Tacke, F.; Wiedenmann, B.; Roderburg, C.; Jann, H. Somatostatin Analogues in the Treatment of Neuroendocrine Tumors: Past, Present and Future. Int. J. Mol. Sci. 2019, 20, 3049. [Google Scholar] [CrossRef]
- Grasso, L.F.; Pivonello, R.; Colao, A. Investigational Therapies for Acromegaly. Expert Opin. Investig. Drugs 2013, 22, 955–963. [Google Scholar] [CrossRef][Green Version]
- Bianchi, A.; Valentini, F.; Iuorio, R.; Poggi, M.; Baldelli, R.; Passeri, M.; Giampietro, A.; Tartaglione, L.; Chiloiro, S.; Appetecchia, M.; et al. Long-Term Treatment of Somatostatin Analog-Refractory Growth Hormone-Secreting Pituitary Tumors with Pegvisomant Alone or Combined with Long-Acting Somatostatin Analogs: A Retrospective Analysis of Clinical Practice and Outcomes. J. Exp. Clin. Cancer Res. 2013, 32, 40. [Google Scholar] [CrossRef]
- Aiello, A.; Cassarino, M.F.; Nanni, S.; Sesta, A.; Ferraú, F.; Grassi, C.; Losa, M.; Trimarchi, F.; Pontecorvi, A.; Cannavò, S.; et al. Establishment of a Protocol to Extend the Lifespan of Human Hormone-Secreting Pituitary Adenoma Cells. Endocrine 2018, 59, 102–108. [Google Scholar] [CrossRef]
- Cui, Y.; Li, C.; Jiang, Z.; Zhang, S.; Li, Q.; Liu, X.; Zhou, Y.; Li, R.; Wei, L.; Li, L.; et al. Single-Cell Transcriptome and Genome Analyses of Pituitary Neuroendocrine Tumors. Neuro. Oncol. 2021, 23, 1859–1871. [Google Scholar] [CrossRef]
- Zhang, Q.; Yao, B.; Long, X.; Chen, Z.; He, M.; Wu, Y.; Qiao, N.; Ma, Z.; Ye, Z.; Zhang, Y.; et al. Single-Cell Sequencing Identifies Differentiation-Related Markers for Molecular Classification and Recurrence Prediction of PitNET. Cell Rep. Med. 2023, 4, 100934. [Google Scholar] [CrossRef]
- Tatsi, C.; Stratakis, C.A. The Genetics of Pituitary Adenomas. Best Pract. Res. Clin. Endocrinol. Metab. 2019, 9, 30. [Google Scholar] [CrossRef]
- Liu, Y.; Zhuang, D.; Hou, R.; Li, J.; Xu, G.; Song, T.; Chen, L.; Yan, G.; Pang, Q.; Zhu, J. Shotgun Proteomic Analysis of Microdissected Postmortem Human Pituitary Using Complementary Two-Dimensional Liquid Chromatography Coupled with Tandem Mass Spectrometer. Anal. Chim. Acta 2011, 688, 183–190. [Google Scholar] [CrossRef]
- Rahimian, N.; Sheida, A.; Rajabi, M.; Heidari, M.M.; Tobeiha, M.; Esfahani, P.V.; Ahmadi Asouri, S.; Hamblin, M.R.; Mohamadzadeh, O.; Motamedzadeh, A.; et al. Noncoding RNAs and Exosomal Noncoding RNAs in Pituitary Adenoma. Pathol. Res. Pract. 2023, 248, 154649. [Google Scholar] [CrossRef]
- Esmaeili Motlagh, P.; Ghafouri-Fard, S.; Eslami, S.; Sharifi, G.; Taheri, M. Expression Assays of Selected lncRNAs in Non-Functioning Pituitary Adenomas. Discov. Oncol. 2024, 15, 486. [Google Scholar] [CrossRef]
- Yin, H.; Zheng, X.; Tang, X.; Zang, Z.; Li, B.; He, S.; Shen, R.; Yang, H.; Li, S. Potential Biomarkers and lncRNA-mRNA Regulatory Networks in Invasive Growth Hormone-Secreting Pituitary Adenomas. J. Endocrinol. Investig. 2021, 44, 1947–1959. [Google Scholar] [CrossRef]
- Catellani, C.; Ravegnini, G.; Sartori, C.; Angelini, S.; Street, M.E. GH and IGF System: The Regulatory Role of miRNAs and lncRNAs in Cancer. Front. Endocrinol. 2021, 12, 701246. [Google Scholar] [CrossRef]
- Lu, T.; Yu, C.; Ni, H.; Liang, W.; Yan, H.; Jin, W. Expression of the Long Noncoding RNA H19 and MALAT-1 in Growth Hormone-secreting Pituitary Adenomas and Its Relationship to Tumor Behavior. Int. J. Dev. Neurosci. 2018, 67, 46–50. [Google Scholar] [CrossRef]
- Babraham Bioinformatics-FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 2 February 2024).
- Martin, M. CUTADAPT Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread Aligner: Fast, Accurate and Scalable Read Mapping by Seed-and-Vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- R Core Team. R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 6 February 2024).
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Possieri, C.; Locantore, P.; Salis, C.; Bacci, L.; Aiello, A.; Fadda, G.; De Crea, C.; Raffaelli, M.; Bellantone, R.; Grassi, C.; et al. Combined Molecular and Mathematical Analysis of Long Noncoding RNAs Expression in Fine Needle Aspiration Biopsies as Novel Tool for Early Diagnosis of Thyroid Cancer. Endocrine 2021, 72, 711–720. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, H.; Liu, Q.; Xie, W.; Li, B.; Cheng, S.; Guo, J.; Fang, Q.; Zhu, H.; Wang, Z.; et al. Functional Characterization of DLK1/MEG3 Locus on Chromosome 14q32.2 Reveals the Differentiation of Pituitary Neuroendocrine Tumors. Aging 2021, 13, 1422–1439. [Google Scholar] [CrossRef]
- Piña, J.O.; Faucz, F.R.; Padilla, C.; Floudas, C.S.; Chittiboina, P.; Quezado, M.; Tatsi, C. Spatial Transcriptomic Analysis of Pituitary Corticotroph Tumors Unveils Intratumor Heterogeneity. medRxiv 2023. [Google Scholar] [CrossRef]
- Beylerli, O.; Beeraka, N.M.; Gareev, I.; Pavlov, V.; Yang, G.; Liang, Y.; Aliev, G. MiRNAs as Noninvasive Biomarkers and Therapeutic Agents of Pituitary Adenomas. Int. J. Mol. Sci. 2020, 21, 7287. [Google Scholar] [CrossRef]
- Du, Q.; Yao, D.-S.; Wang, Y.-W.; Cheng, C. Research Progress on lncRNA Functions and Mechanisms in Pituitary Adenomas. Horm. Metab. Res. 2020, 52, 280–288. [Google Scholar] [CrossRef]
- Fu, D.; Zhang, Y.; Cui, H. Long Noncoding RNA CCAT2 Is Activated by E2F1 and Exerts Oncogenic Properties by Interacting with PTTG1 in Pituitary Adenomas. Am. J. Cancer Res. 2018, 8, 245–255. [Google Scholar] [PubMed]
- Wu, Z.; Zheng, Y.; Xie, W.; Li, Q.; Zhang, Y.; Ren, B.; Cai, L.; Cheng, Y.; Tang, H.; Su, Z.; et al. The Long Noncoding RNA-H19/miRNA-93a/ATG7 Axis Regulates the Sensitivity of Pituitary Adenomas to Dopamine Agonists. Mol. Cell Endocrinol. 2020, 518, 111033. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Safarzadeh, A.; Hussen, B.M.; Taheri, M.; Eghbali, A. Expression of LINC00174 in Different Cancers: Review of the Literature and Bioinformatics Analyses. Pathol. Res. Pract. 2023, 248, 154617. [Google Scholar] [CrossRef] [PubMed]
| MALAT1 | NEAT1 | MEG3 | H19 | |
|---|---|---|---|---|
| Patient | MEAN ± SD. | MEAN ± SD. | MEAN ± SD. | MEAN ± SD. |
| A | 41,327.45 ± 7845.48 | 631.04 ± 120.14 | 242.14 ± 82.49 | 1.12 ± 0.58 |
| B | 60,690.38 ± 1508.89 | 299.37 ± 61.83 | 221.65 ± 88.91 | 0.41 ± 0.28 |
| C | 62,887.38 ± 15,820.38 | 745.55 ± 169.56 | 160.94 ± 40.72 | 1.99 ± 0.79 |
| D | 24,357.52 ± 3778.03 | 744.11 ± 151.57 | 175.66 ± 22.30 | 4.11 ± 1.18 |
| E | 131,203.57 ± 97,292.85 | 2733.41 ± 1732.73 | 606.04 ± 392.20 | 1.92 ± 1.64 |
| F | 266,572.33 ± 30,774.71 | 2450.00 ± 310.98 | 6091.61 ± 835.53 | 2.82 ± 0.93 |
| G | 267,926.36 ± 33,573.87 | 2222.51 ± 334.64 | 1855.07 ± 191.66 | 5.79 ± 3.39 |
| H | 8164.74 ± 652.60 | 991.11 ± 147.02 | 473.25 ± 30.93 | 1.48 ± 0.26 |
| I | 149,113.10 ± 5143.36 | 2030.48 ± 84.36 | 1876.73 ± 215.19 | 5.86 ± 1.98 |
| L | 247,521.69 ± 109,943.04 | 2345.05 ± 851.72 | 1810.35 ± 685.98 | 3.54 ± 3.25 |
| M | 68,399.03 ± 22,877.46 | 379.42 ± 103.98 | 213.82 ± 60.33 | 0.67 ± 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cis, L.; Nanni, S.; Gessi, M.; Bianchi, A.; De Martino, S.; Pecci, V.; Bonvissuto, D.; Carlino, A.; Giacò, L.; Rindi, G.; et al. LCM-RNAseq Highlights Intratumor Heterogeneity and a lncRNA Signature from Archival Tissues of GH-Secreting PitNETs. Genes 2024, 15, 1426. https://doi.org/10.3390/genes15111426
Cis L, Nanni S, Gessi M, Bianchi A, De Martino S, Pecci V, Bonvissuto D, Carlino A, Giacò L, Rindi G, et al. LCM-RNAseq Highlights Intratumor Heterogeneity and a lncRNA Signature from Archival Tissues of GH-Secreting PitNETs. Genes. 2024; 15(11):1426. https://doi.org/10.3390/genes15111426
Chicago/Turabian StyleCis, Luca, Simona Nanni, Marco Gessi, Antonio Bianchi, Sara De Martino, Valeria Pecci, Davide Bonvissuto, Angela Carlino, Luciano Giacò, Guido Rindi, and et al. 2024. "LCM-RNAseq Highlights Intratumor Heterogeneity and a lncRNA Signature from Archival Tissues of GH-Secreting PitNETs" Genes 15, no. 11: 1426. https://doi.org/10.3390/genes15111426
APA StyleCis, L., Nanni, S., Gessi, M., Bianchi, A., De Martino, S., Pecci, V., Bonvissuto, D., Carlino, A., Giacò, L., Rindi, G., Sette, C., Grassi, C., Gaetano, C., Pontecorvi, A., & Farsetti, A. (2024). LCM-RNAseq Highlights Intratumor Heterogeneity and a lncRNA Signature from Archival Tissues of GH-Secreting PitNETs. Genes, 15(11), 1426. https://doi.org/10.3390/genes15111426









