RNAi Knockdown of EHMT2 in Maternal Expression of Prader–Willi Syndrome Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Procedures
2.2. Genotyping
2.3. Cell Culture
2.3.1. iPSC Lines
2.3.2. iPSC-Derived Neuronal Induction
2.3.3. Mouse Primary Neurons
2.4. siRNA Design
2.5. Plasmid Construct
2.6. Cell Transfection with siRNA
2.7. Cell Transfection with Plasmid
2.8. Transduction of iPSC-Derived Cells with Lentivirus
2.9. Quantitative Real-Time Reverse Transcription (qRT)-PCR
2.10. Western Blot Analysis
Statistics
3. Results
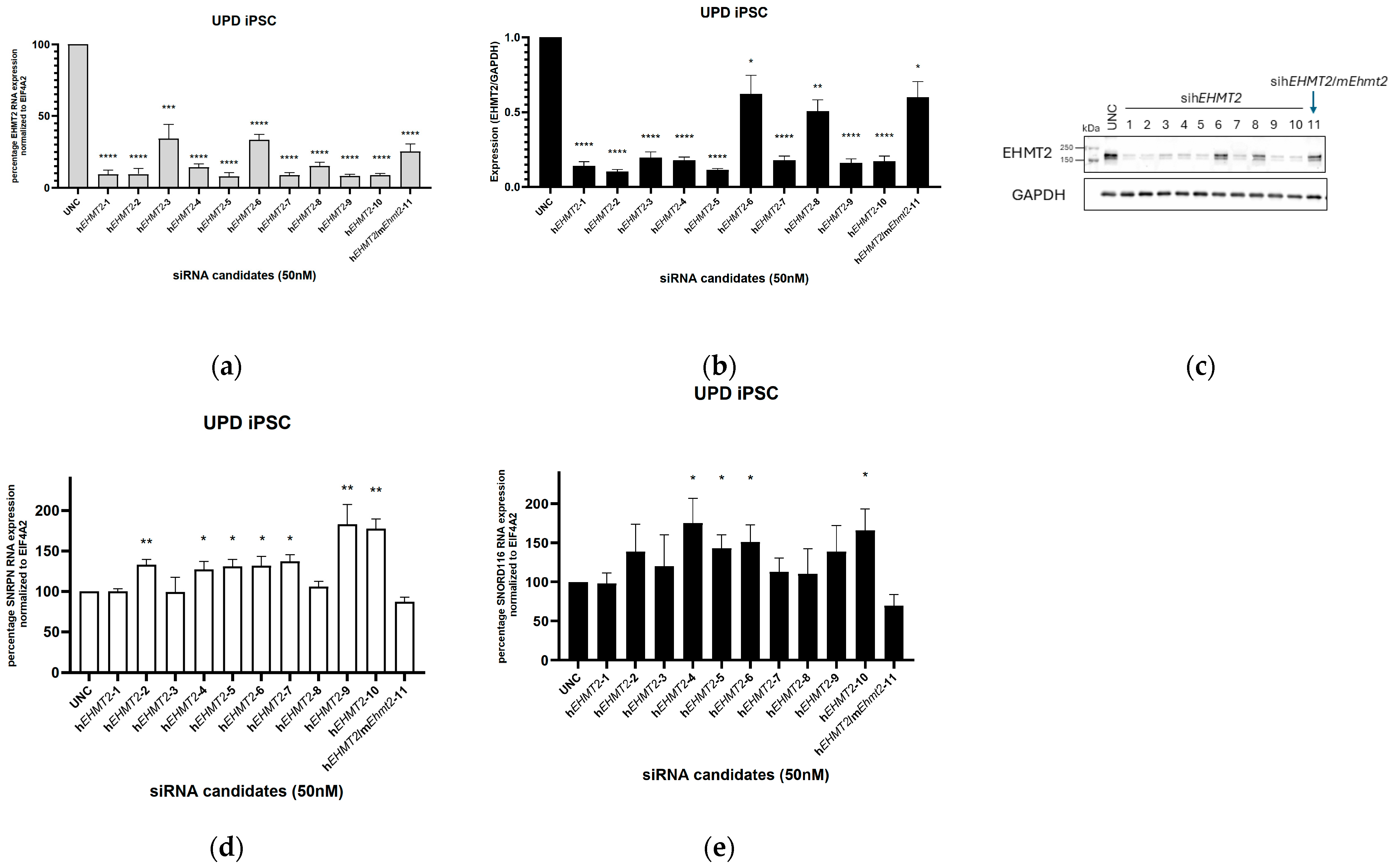
3.1. siRNA Knockdown of EHMT2 in PWS-UPD iPSCs
3.2. siRNA Knockdown of Ehmt2 in Primary Mouse Neurons
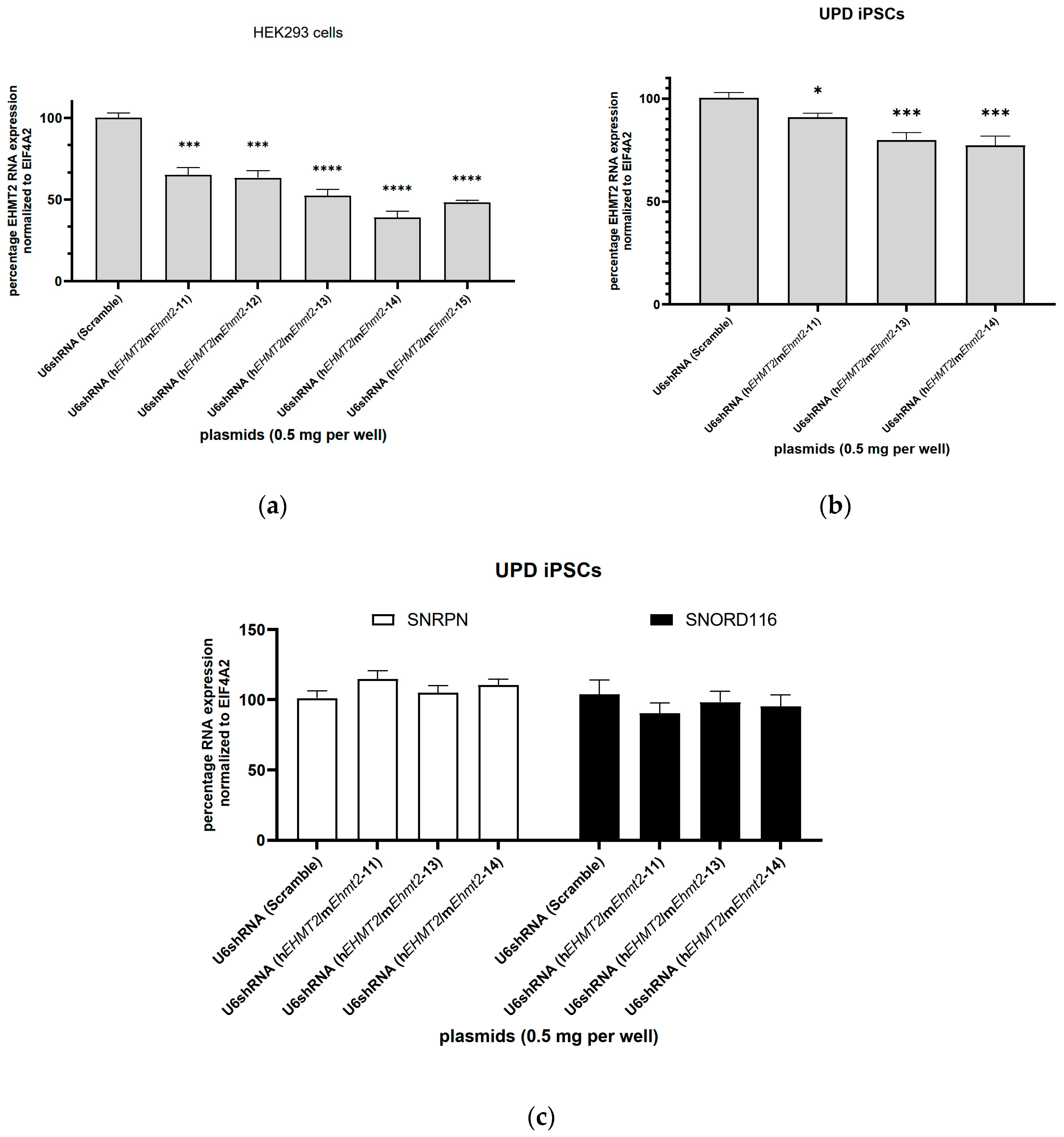
3.3. shRNA Knockdown of EHMT2 in HEK293 Cells
3.4. shRNA Knockdown of EHMT2 in PWS-iPSCs
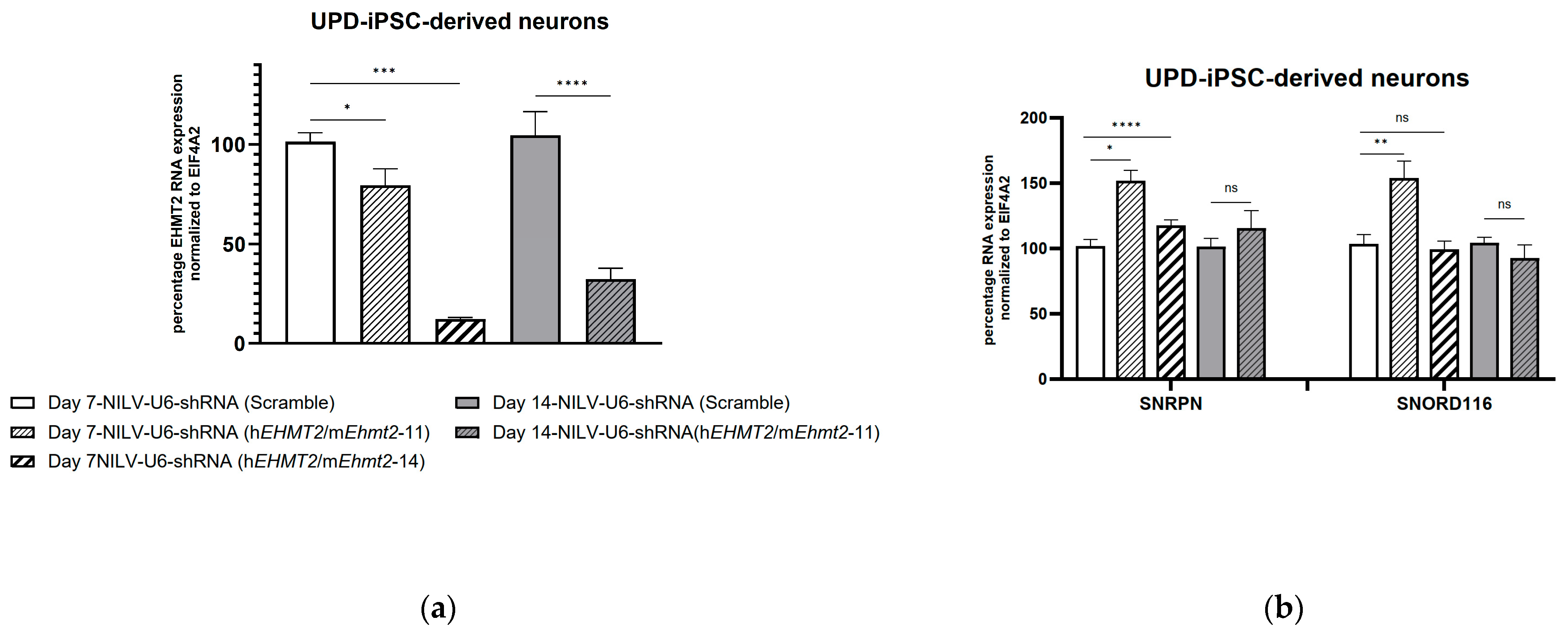
3.5. shRNA Knockdown of EHMT2 in PWS-iPSC-Derived Neurons 7 Days and 14 Days Post Transduction with Lentiviral Constructs
3.6. shRNA Knockdown of EHMT2 in PWS 2-9-iPSC-Derived Neurons 21 Days Post Transduction with Lentiviral Constructs
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Butler, M.G.; Miller, J.L.; Forster, J.L. Prader-Willi Syndrome—Clinical Genetics, Diagnosis and Treatment Approaches: An Update. Curr. Pediatr. Rev. 2019, 15, 207–244. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, S.B.; Schwartz, S.; Miller, J.L.; Driscoll, D.J. Prader-Willi syndrome. Genet. Med. 2012, 14, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Godler, D.E.; Butler, M.G. Special Issue: Genetics of Prader-Willi Syndrome. Genes 2021, 12, 1429. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G.; Hartin, S.N.; Hossain, W.A.; Manzardo, A.M.; Kimonis, V.; Dykens, E.; Gold, J.A.; Kim, S.J.; Weisensel, N.; Tamura, R.; et al. Molecular genetic classification in Prader-Willi syndrome: A multisite cohort study. J. Med. Genet. 2019, 56, 149–153. [Google Scholar] [CrossRef]
- Cassidy, S.B.; Dykens, E.; Williams, C.A. Prader-Willi and Angelman syndromes: Sister imprinted disorders. Am. J. Med. Genet. 2000, 97, 136–146. [Google Scholar] [CrossRef]
- Buiting, K.; Dittrich, B.; Robinson, W.P.; Guitart, M.; Abeliovich, D.; Lerer, I.; Horsthemke, B. Detection of aberrant DNA methylation in unique Prader-Willi syndrome patients and its diagnostic implications. Hum. Mol. Genet. 1994, 3, 893–895. [Google Scholar] [CrossRef][Green Version]
- de Smith, A.J.; Purmann, C.; Walters, R.G.; Ellis, R.J.; Holder, S.E.; Van Haelst, M.M.; Brady, A.F.; Fairbrother, U.L.; Dattani, M.; Keogh, J.M.; et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 2009, 18, 3257–3265. [Google Scholar] [CrossRef]
- Sahoo, T.; del Gaudio, D.; German, J.R.; Shinawi, M.; Peters, S.U.; Person, R.E.; Garnica, A.; Cheung, S.W.; Beaudet, A.L. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008, 40, 719–721. [Google Scholar] [CrossRef]
- Schulze, A.; Hansen, C.; Skakkebaek, N.E.; Brondum-Nielsen, K.; Ledbeter, D.H.; Tommerup, N. Exclusion of SNRPN as a major determinant of Prader-Willi syndrome by a translocation breakpoint. Nat. Genet. 1996, 12, 452–454. [Google Scholar] [CrossRef]
- Duker, A.L.; Ballif, B.C.; Bawle, E.V.; Person, R.E.; Mahadevan, S.; Alliman, S.; Thompson, R.; Traylor, R.; Bejjani, B.A.; Shaffer, L.G.; et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur. J. Hum. Genet. 2010, 18, 1196–1201. [Google Scholar] [CrossRef]
- Grootjen, L.N.; Juriaans, A.F.; Kerkhof, G.F.; Hokken-Koelega, A.C.S. Atypical 15q11.2-q13 Deletions and the Prader-Willi Phenotype. J. Clin. Med. 2022, 11, 4636. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; AlHumaidi, S.S.; Faqeih, E.A.; Pitel, B.A.; Lundquist, P.; Aypar, U. A novel deletion of SNURF/SNRPN exon 1 in a patient with Prader-Willi-like phenotype. Eur. J. Med. Genet. 2017, 60, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Crenshaw, M.M.; Graw, S.L.; Slavov, D.; Boyle, T.A.; Pique, D.G.; Taylor, M.; Baker, P., II. An Atypical 15q11.2 Microdeletion Not Involving SNORD116 Resulting in Prader-Willi Syndrome. Case Rep. Genet. 2023, 2023, 4225092. [Google Scholar] [CrossRef]
- Saitoh, S.; Wada, T. Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader-Willi syndrome. Am. J. Hum. Genet. 2000, 66, 1958–1962. [Google Scholar] [CrossRef]
- Fulmer-Smentek, S.B.; Francke, U. Association of acetylated histones with paternally expressed genes in the Prader-Willi deletion region. Hum. Mol. Genet. 2001, 10, 645–652. [Google Scholar] [CrossRef]
- Cruvinel, E.; Budinetz, T.; Germain, N.; Chamberlain, S.; Lalande, M.; Martins-Taylor, K. Reactivation of maternal SNORD116 cluster via SETDB1 knockdown in Prader-Willi syndrome iPSCs. Hum. Mol. Genet. 2014, 23, 4674–4685. [Google Scholar] [CrossRef] [PubMed]
- Recillas-Targa, F. DNA methylation, chromatin boundaries, and mechanisms of genomic imprinting. Arch. Med. Res. 2002, 33, 428–438. [Google Scholar] [CrossRef]
- Kacem, S.; Feil, R. Chromatin mechanisms in genomic imprinting. Mamm. Genome 2009, 20, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T. The epigenetic magic of histone lysine methylation. FEBS J. 2006, 273, 3121–3135. [Google Scholar] [CrossRef]
- Zhang, R.H.; Judson, R.N.; Liu, D.Y.; Kast, J.; Rossi, F.M. The lysine methyltransferase Ehmt2/G9a is dispensable for skeletal muscle development and regeneration. Skelet. Muscle 2016, 6, 22. [Google Scholar] [CrossRef]
- Liang, L.; Gu, X.; Zhao, J.Y.; Wu, S.; Miao, X.; Xiao, J.; Mo, K.; Zhang, J.; Lutz, B.M.; Bekker, A.; et al. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci. Rep. 2016, 6, 37704. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Du, S.; Wang, K.; Guo, X.; Mao, Q.; Feng, X.; Huang, L.; Wu, S.; Hou, B.; Chang, Y.J.; et al. Downregulation of a Dorsal Root Ganglion-Specifically Enriched Long Noncoding RNA is Required for Neuropathic Pain by Negatively Regulating RALY-Triggered Ehmt2 Expression. Adv. Sci. 2021, 8, e2004515. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.; Kosowan, J.; Samsom, J.; Leung, L.; Zhang, K.L.; Li, Y.X.; Xiong, Y.; Jin, J.; Petronis, A.; Oh, G.; et al. Inhibition of the G9a/GLP histone methyltransferase complex modulates anxiety-related behavior in mice. Acta Pharmacol. Sin. 2018, 39, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, H.M.; Xiong, Y.; Sciaky, N.; Hulbert, S.W.; Cao, X.; Everitt, J.I.; Jin, J.; Roth, B.L.; Jiang, Y.H. Targeting the histone methyltransferase G9a activates imprinted genes and improves survival of a mouse model of Prader-Willi syndrome. Nat. Med. 2017, 23, 213–222. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Rand, T.A.; Ginalski, K.; Grishin, N.V.; Wang, X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. USA 2004, 101, 14385–14389. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef]
- Corydon, I.J.; Fabian-Jessing, B.K.; Jakobsen, T.S.; Jorgensen, A.C.; Jensen, E.G.; Askou, A.L.; Aagaard, L.; Corydon, T.J. 25 years of maturation: A systematic review of RNAi in the clinic. Mol. Ther. Nucleic Acids 2023, 33, 469–482. [Google Scholar] [CrossRef]
- Ding, F.; Prints, Y.; Dhar, M.S.; Johnson, D.K.; Garnacho-Montero, C.; Nicholls, R.D.; Francke, U. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm. Genome 2005, 16, 424–431. [Google Scholar] [CrossRef]
- Osborne-Lawrence, S.; Lawrence, C.; Metzger, N.P.; Klavon, J.; Baig, H.R.; Richard, C.; Varshney, S.; Gupta, D.; Singh, O.; Ogden, S.B.; et al. Effects of thermoneutrality on food intake, body weight, and body composition in a Prader-Willi syndrome mouse model. Obesity 2023, 31, 1644–1654. [Google Scholar] [CrossRef]
- Martins-Taylor, K.; Hsiao, J.S.; Chen, P.F.; Glatt-Deeley, H.; De Smith, A.J.; Blakemore, A.I.; Lalande, M.; Chamberlain, S.J. Imprinted expression of UBE3A in non-neuronal cells from a Prader-Willi syndrome patient with an atypical deletion. Hum. Mol. Genet. 2014, 23, 2364–2373. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, G.M., III; Lee, S.H.; Singh, D.; Yuan, Y.; Ng, Y.G.; Reichardt, L.F.; Arikkath, J. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 2012, 7, 1741–1754. [Google Scholar] [CrossRef] [PubMed]
- Moutin, E.; Hemonnot, A.L.; Seube, V.; Linck, N.; Rassendren, F.; Perroy, J.; Compan, V. Procedures for Culturing and Genetically Manipulating Murine Hippocampal Postnatal Neurons. Front. Synaptic Neurosci. 2020, 12, 19. [Google Scholar] [CrossRef]
- Boudreau, R.L.; Spengler, R.M.; Hylock, R.H.; Kusenda, B.J.; Davis, H.A.; Eichmann, D.A.; Davidson, B.L. siSPOTR: A tool for designing highly specific and potent siRNAs for human and mouse. Nucleic Acids Res. 2013, 41, e9. [Google Scholar] [CrossRef]
- Langouet, M.; Gorka, D.; Orniacki, C.; Dupont-Thibert, C.M.; Chung, M.S.; Glatt-Deeley, H.R.; Germain, N.; Crandall, L.J.; Cotney, J.L.; Stoddard, C.E.; et al. Specific ZNF274 binding interference at SNORD116 activates the maternal transcripts in Prader-Willi syndrome neurons. Hum. Mol. Genet. 2020, 29, 3285–3295. [Google Scholar] [CrossRef]
- Cavaille, J.; Buiting, K.; Kiefmann, M.; Lalande, M.; Brannan, C.I.; Horsthemke, B.; Bachellerie, J.P.; Brosius, J.; Huttenhofer, A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 2000, 97, 14311–14316. [Google Scholar] [CrossRef]
- Yang, T.; Adamson, T.E.; Resnick, J.L.; Leff, S.; Wevrick, R.; Francke, U.; Jenkins, N.A.; Copeland, N.G.; Brannan, C.I. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat. Genet. 1998, 19, 25–31. [Google Scholar] [CrossRef]
- Langouet, M.; Glatt-Deeley, H.R.; Chung, M.S.; Dupont-Thibert, C.M.; Mathieux, E.; Banda, E.C.; Stoddard, C.E.; Crandall, L.; Lalande, M. Zinc finger protein 274 regulates imprinted expression of transcripts in Prader-Willi syndrome neurons. Hum. Mol. Genet. 2018, 27, 505–515. [Google Scholar] [CrossRef]
- Wilson, C.; Giono, L.E.; Rozes-Salvador, V.; Fiszbein, A.; Kornblihtt, A.R.; Caceres, A. The Histone Methyltransferase G9a Controls Axon Growth by Targeting the RhoA Signaling Pathway. Cell Rep. 2020, 31, 107639. [Google Scholar] [CrossRef]
- Barral, A.; Pozo, G.; Ducrot, L.; Papadopoulos, G.L.; Sauzet, S.; Oldfield, A.J.; Cavalli, G.; Dejardin, J. SETDB1/NSD-dependent H3K9me3/H3K36me3 dual heterochromatin maintains gene expression profiles by bookmarking poised enhancers. Mol. Cell 2022, 82, 816–832.e12. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.L.F.; Cheung, M.F.; Chan, L.Y.; Leung, D. Cell-type differential targeting of SETDB1 prevents aberrant CTCF binding, chromatin looping, and cis-regulatory interactions. Nat. Commun. 2024, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Weirich, S.; Khella, M.S.; Jeltsch, A. Structure, Activity and Function of the Suv39h1 and Suv39h2 Protein Lysine Methyltransferases. Life 2021, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.E.; Northrop, J.P.; Horton, J.R.; Lee, D.Y.; Zhang, X.; Stallcup, M.R.; Cheng, X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 2008, 15, 245–250. [Google Scholar] [CrossRef]
- Tachibana, M.; Sugimoto, K.; Fukushima, T.; Shinkai, Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001, 276, 25309–25317. [Google Scholar] [CrossRef]
- Able, A.A.; Richard, A.J.; Stephens, J.M. TNFalpha Effects on Adipocytes Are Influenced by the Presence of Lysine Methyltransferases, G9a (EHMT2) and GLP (EHMT1). Biology 2023, 12, 674. [Google Scholar] [CrossRef]


| iPSC Lines | Description | Reference | Abbreviation |
|---|---|---|---|
| PWS UPD 1–2 | PWS uniparental disomy | [31] | UPD iPSC |
| PWS 2–9 | PWS small deletion | [31] | SD iPSC |
| shRNA Name | Sequence (5′-3′)—Sense | Sequence (5′-3′)—Antisense | Species Target EHMT2 |
|---|---|---|---|
| hEHMT2/mEhmt2-11 | TAAATGTTGGGTTTGGTAATA | TATTACCAAACCCAACATTTA | hsa and mmu |
| hEHMT2/mEhmt2-12 | GGCGCAAGGCCAAGAAGAAAT | ATTTCTTCTTGGCCTTGCGCC | hsa and mmu |
| hEHMT2/mEhmt2-13 | GCGCAAGGCCAAGAAGAAATG | CATTTCTTCTTGGCCTTGCGC | hsa and mmu |
| hEHMT2/mEhmt2-14 | TGATGTGAGAGAGGATGATTC | GAATCATCCTCTCTCACATCA | hsa and mmu |
| hEHMT2/mEhmt2-15 | GTGAGAGAGGATGATTCTTAC | GTAAGAATCATCCTCTCTCAC | hsa and mmu |
| Primer Name | Sequence | Species | SybR or with Probe |
|---|---|---|---|
| EIF4A2 F | CAACGTGCATTGTGCTTCTT | hsa | SybR |
| EIF4A2 R | ACGACTAACGTCGCTTTGCT | hsa | SybR |
| SNORD116 F | GGATCGATGATGAGTCCCC | hsa | SybR |
| SNORD116 R | TCCGATGAGAACGACGGTAT | hsa | SybR |
| SNRPN F | TTGCTGCGACTGCCAGTATT | hsa | SybR |
| SNRPN R | GCCCATGGGTGGTCTCATAC | hsa | SybR |
| MAGEL2 F | ATCTGGAAGCCCAAGAGGAC | hsa | SybR |
| MAGEL2 R | ACCTGGATAGGGCTTTGGAC | hsa | SybR |
| EHMT2 F | ATGCAGTGGACAAACAGCAG | hsa | SybR |
| EHMT2 R | ACCGTCCTCCTCCTTGCTAT | hsa | SybR |
| Snord116 HG F | TGTTGCTGACTTGCCCTAG | mmu | probe |
| Snord116 HG R | GTTCGATGGAGACTCAGTTGG | mmu | probe |
| mSnord116 (probe) | AAACATGCAGAGGAAATGGCCCC | mmu | probe |
| Eif4 | Mm01730183_gH/4448484 | mmu | probe |
| Ehmt2 F | TGCGTACTCTGTGGATGAGC | mmu | SybR |
| Ehmt2 R | TCTGACTGATTGCCCGACTC | mmu | SybR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaric, V.; Kang, H.R.; Rybalchenko, V.; Zigman, J.M.; Gray, S.J.; Butler, R.K. RNAi Knockdown of EHMT2 in Maternal Expression of Prader–Willi Syndrome Genes. Genes 2024, 15, 1366. https://doi.org/10.3390/genes15111366
Zaric V, Kang HR, Rybalchenko V, Zigman JM, Gray SJ, Butler RK. RNAi Knockdown of EHMT2 in Maternal Expression of Prader–Willi Syndrome Genes. Genes. 2024; 15(11):1366. https://doi.org/10.3390/genes15111366
Chicago/Turabian StyleZaric, Violeta, Hye Ri Kang, Volodymyr Rybalchenko, Jeffrey M. Zigman, Steven J. Gray, and Ryan K. Butler. 2024. "RNAi Knockdown of EHMT2 in Maternal Expression of Prader–Willi Syndrome Genes" Genes 15, no. 11: 1366. https://doi.org/10.3390/genes15111366
APA StyleZaric, V., Kang, H. R., Rybalchenko, V., Zigman, J. M., Gray, S. J., & Butler, R. K. (2024). RNAi Knockdown of EHMT2 in Maternal Expression of Prader–Willi Syndrome Genes. Genes, 15(11), 1366. https://doi.org/10.3390/genes15111366






