Genome-Wide Identification of the Maize Chitinase Gene Family and Analysis of Its Response to Biotic and Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Family Membership Identification and Chromosome Localisation
2.2. Structural Characterization of Physicochemical Properties, Genes, and Proteins of the Maize Chitinase Gene Family
2.3. Phylogenetic Examination of the Maize Chitinase Gene Family
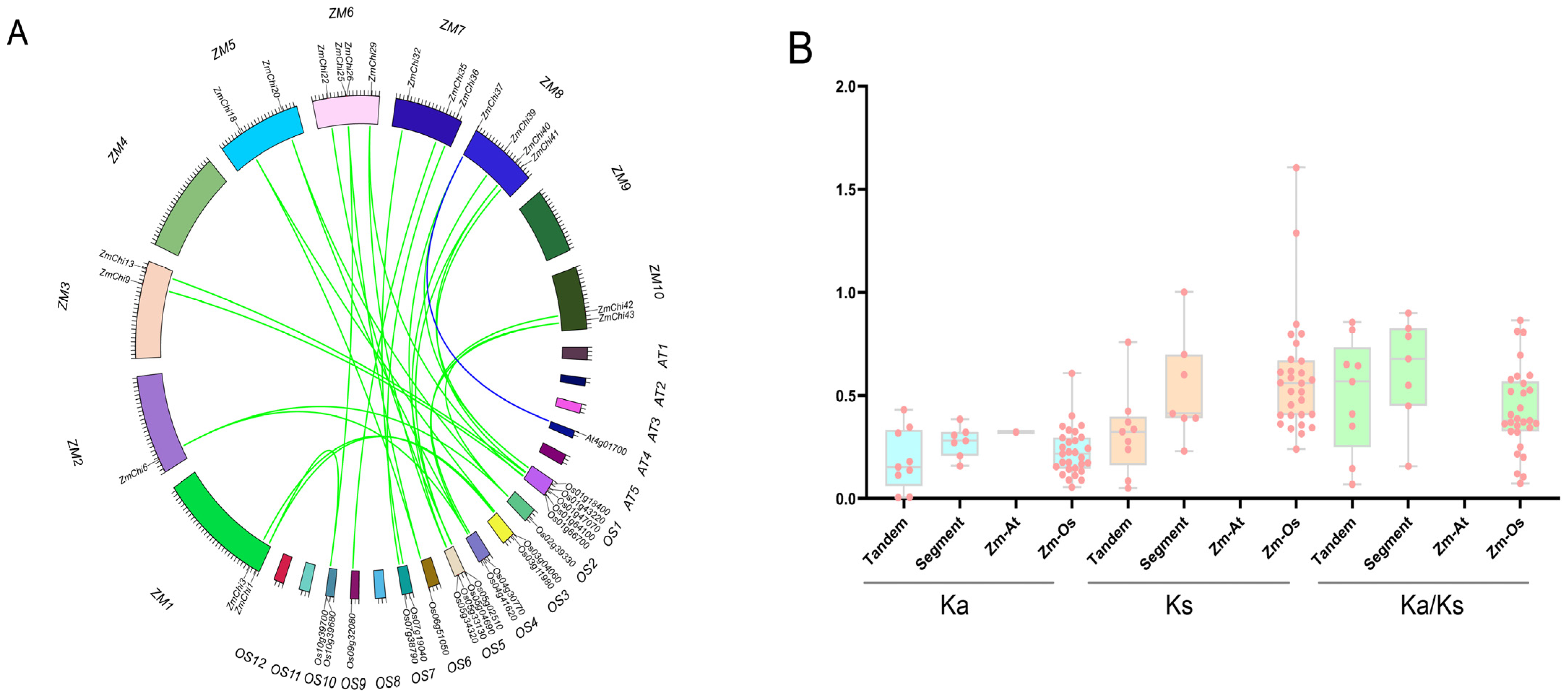
2.4. Chitinase Family Gene Collinearity Analysis in Maize
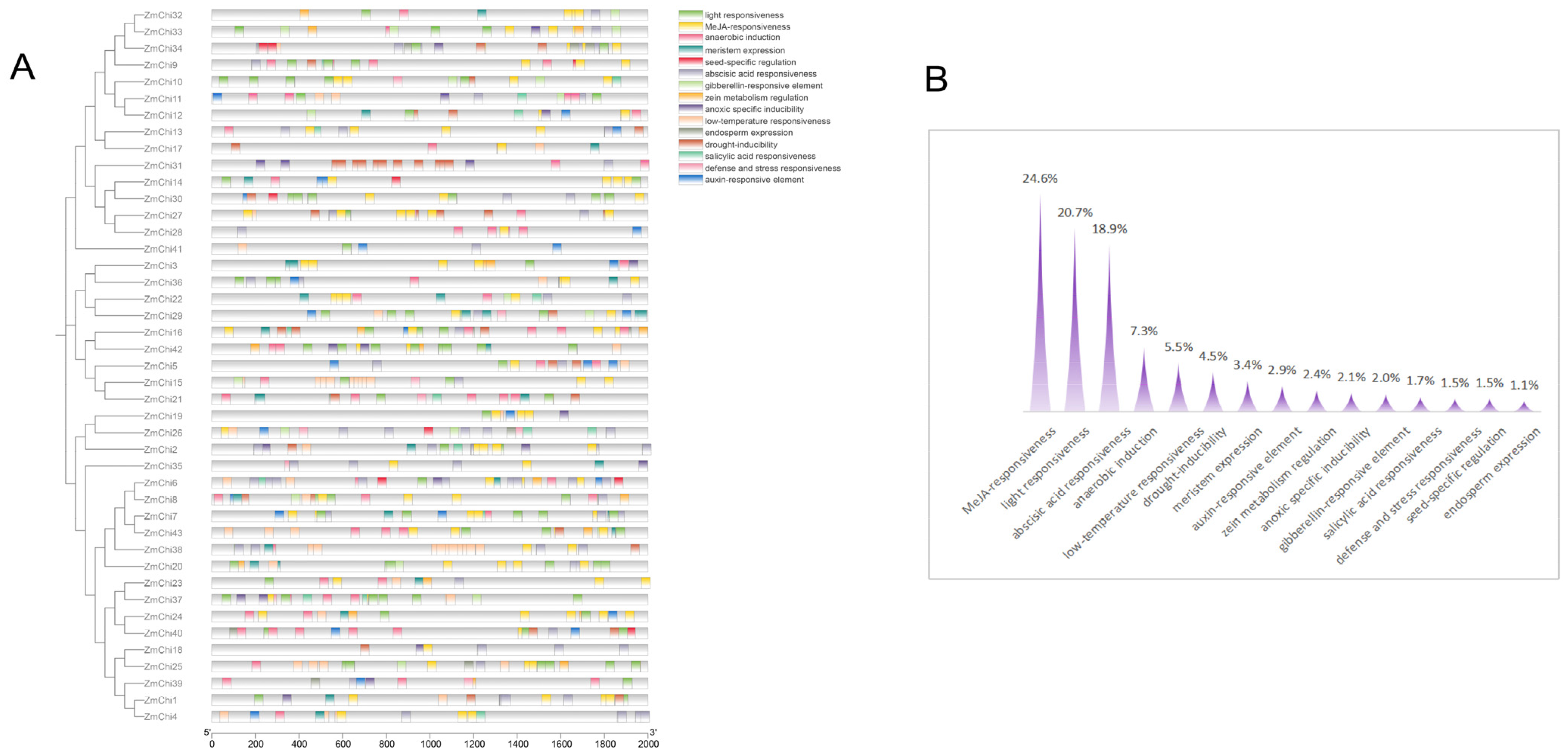
2.5. Cis-Acting Elements and Functional Analyses of Maize Chitinase Genes
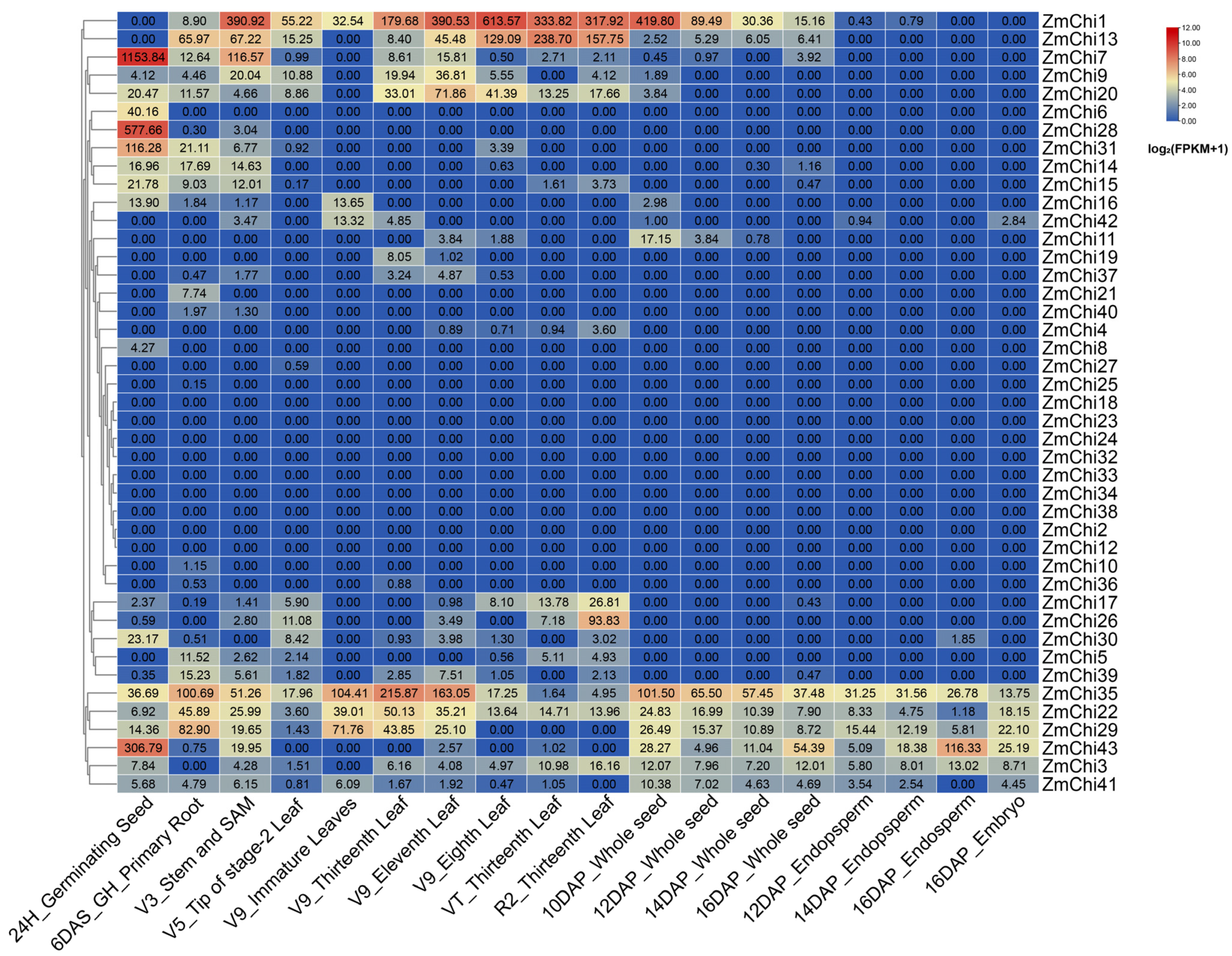
2.6. Tissue-Specific and Stress-Induced Expression Profiles of the Maize Chitinase Gene Family
3. Results
3.1. Basic Information on Members of the Maize Chitinase Gene Family
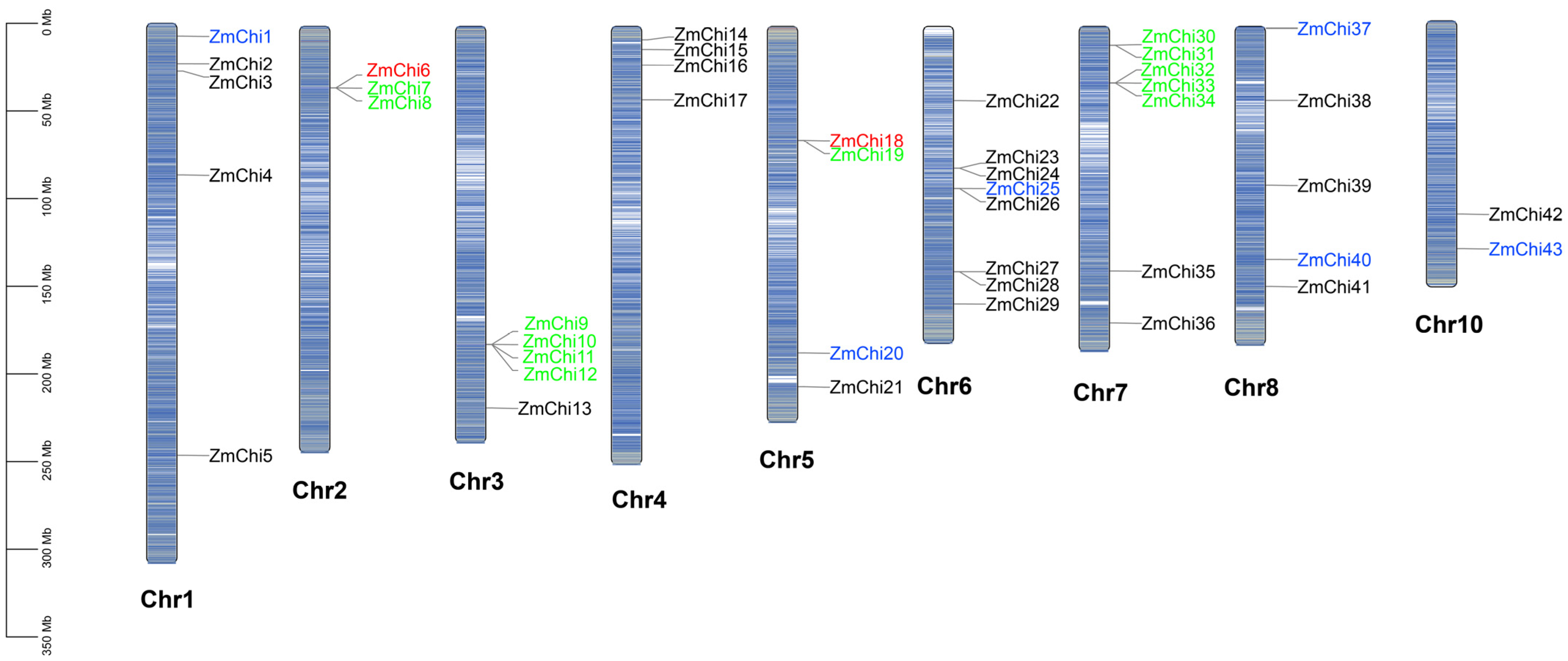
3.2. Chitinase Genes in Maize Are Distributed across Various Chromosomes
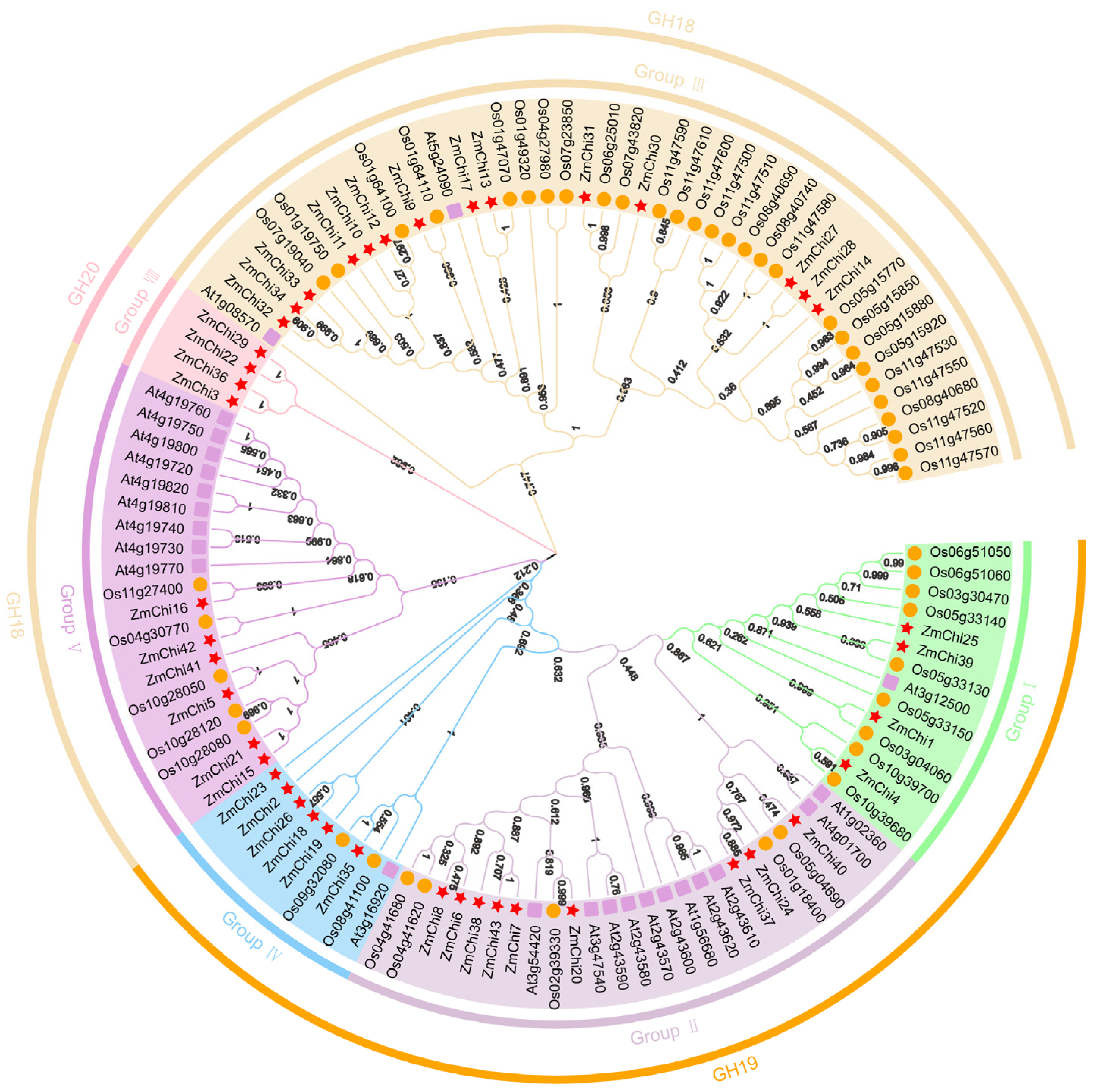
3.3. Phylogenetic Analysis of Maize Chitinase Genes
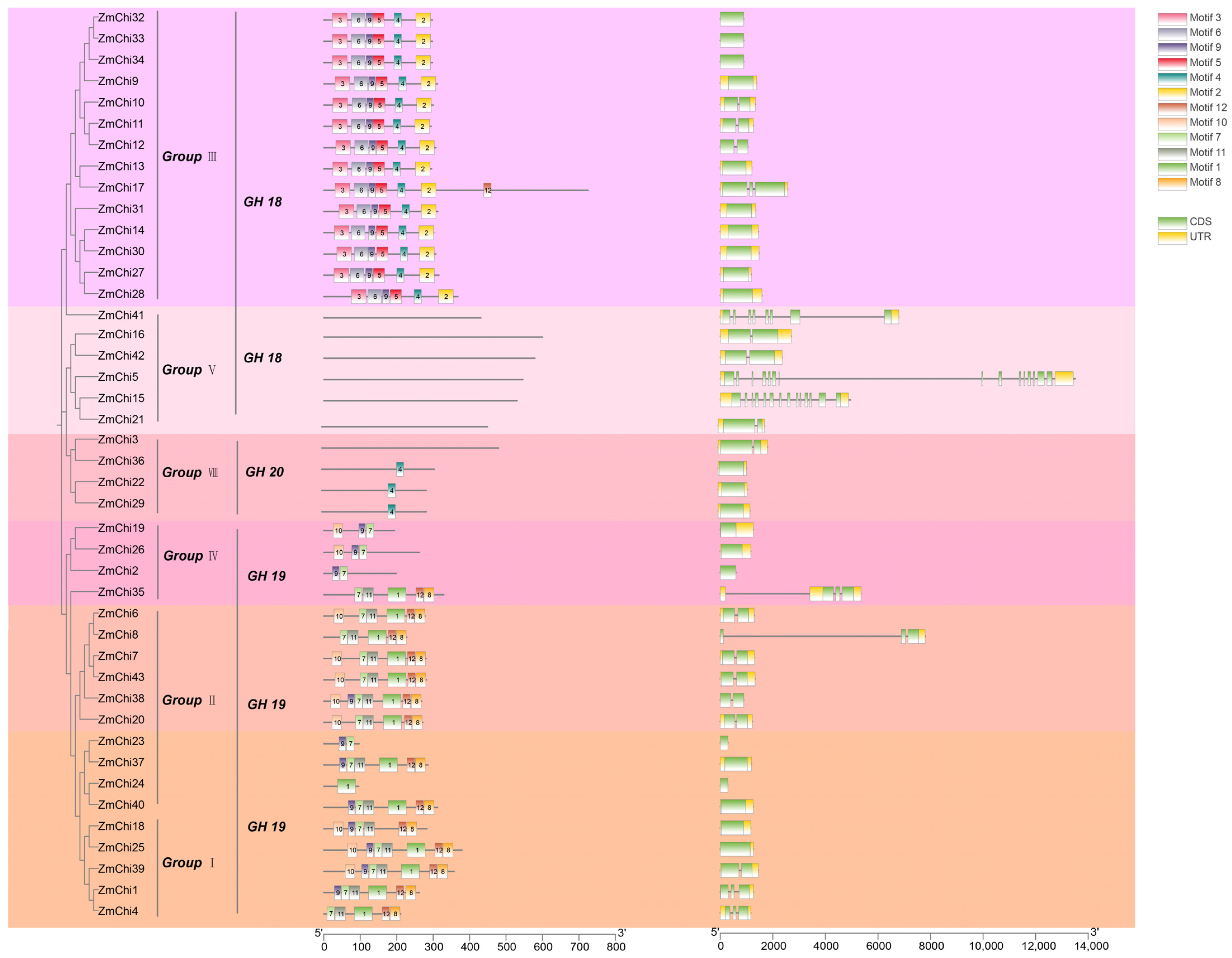
3.4. Gene Structure and Protein Motifs of the Maize Chitinase Gene Family
3.5. Functional Analysis of Chitinase Family Genes in Maize
3.6. Colinearity of Maize Chitinase Family Genes
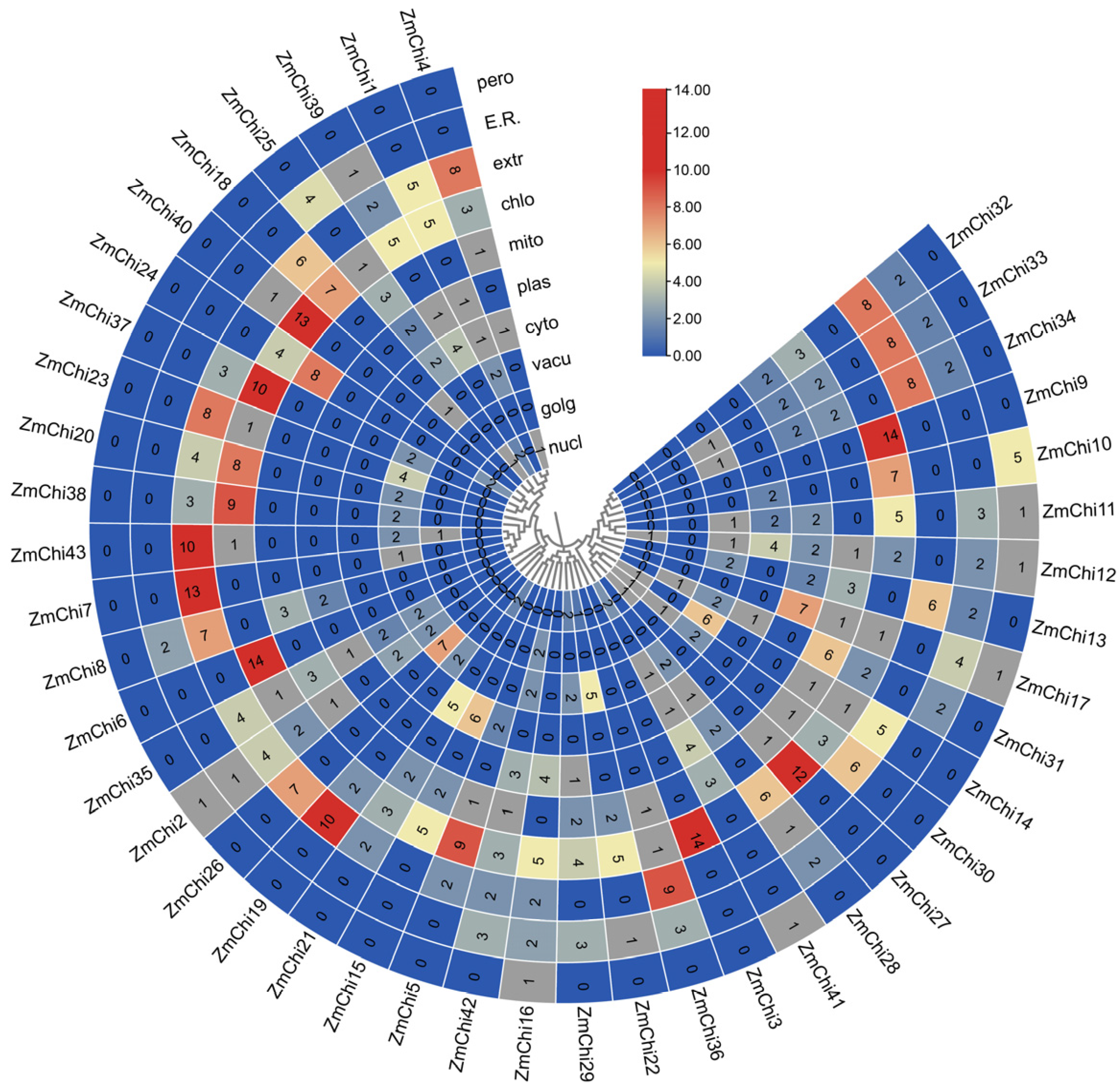
3.7. Tissue-Specific Expression Profiles of Maize Chitinase Genes
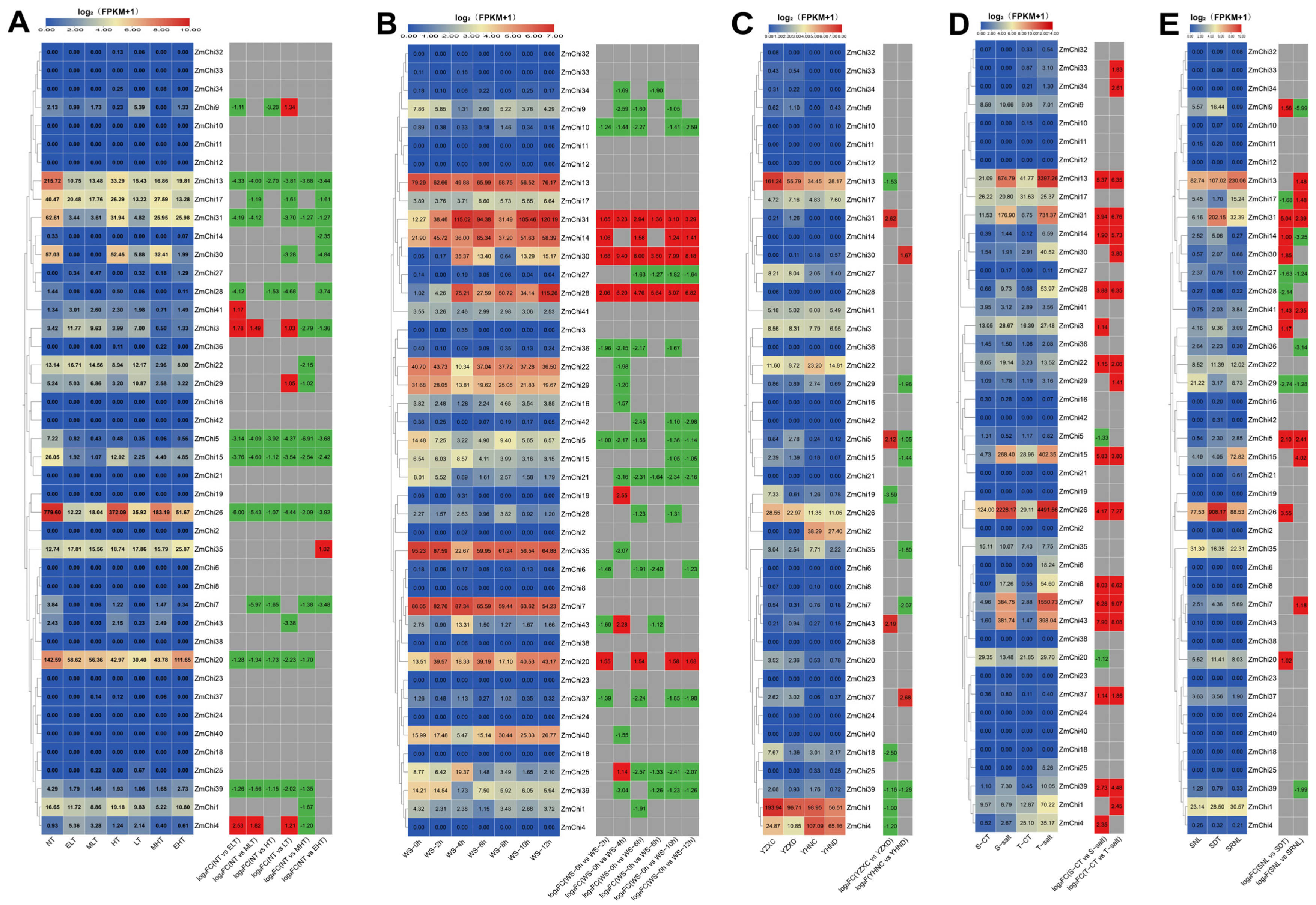
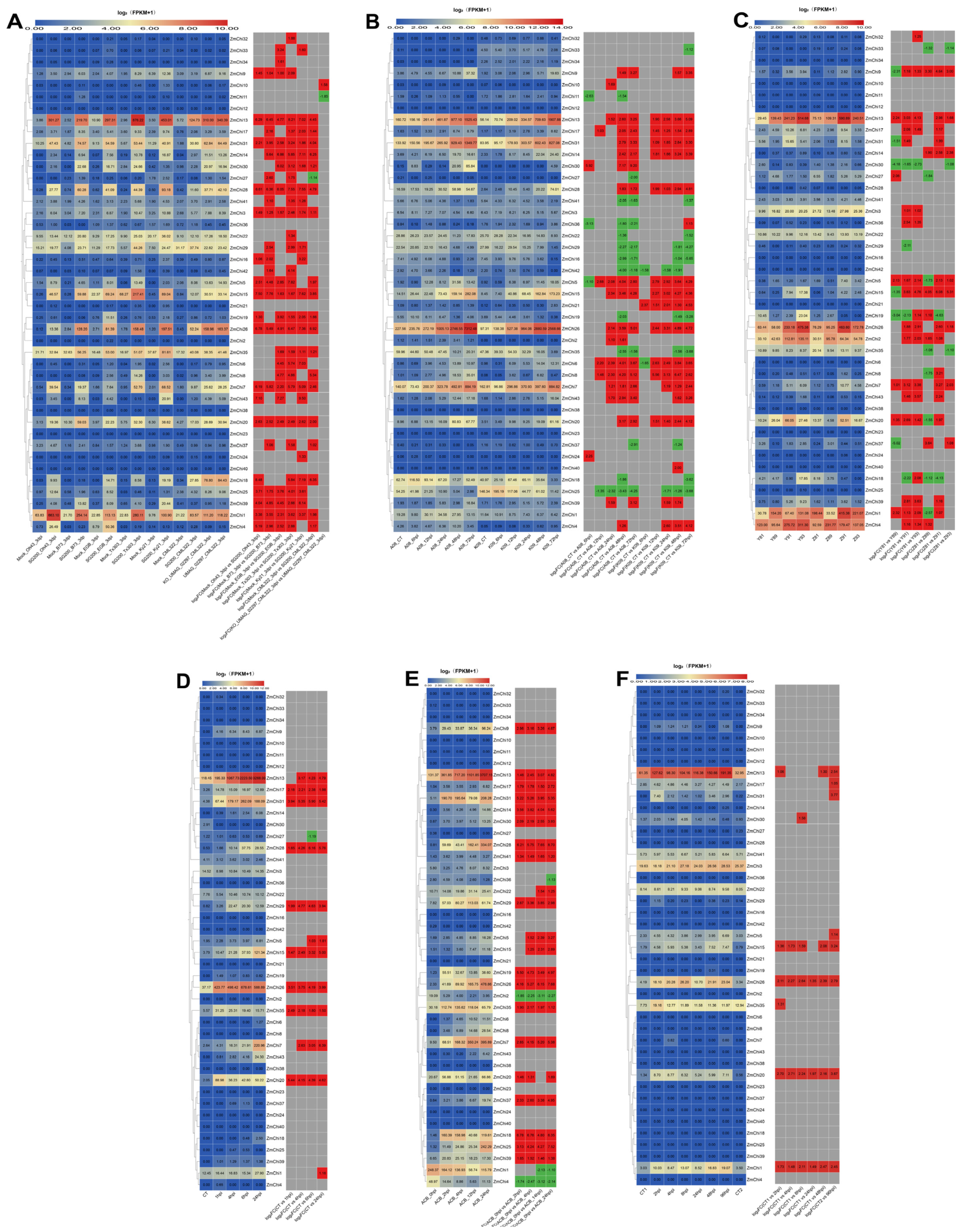
3.8. Analysis of Maize Chitinase Gene Family Expression Profiles under Abiotic Stress
3.9. Analysis of Maize Chitinase Gene Family Expression Profiles under Biotic Stress
3.10. Biological and Abiotic Stress Regulation of Maize Chitinase Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaikh, S.A.; Deshpande, M.V. Chitinolytic enzymes: Their contribution to basic and applied research. World J. Microbiol. Biotechnol. 1993, 9, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Chakdar, H.; Pandiyan, K.; Thapa, S.; Shahid, M.; Singh, A.; Srivastava, A.K.; Saxena, A.K. Bacterial chitinases: Genetics, engineering and applications. World J. Microbiol. Biotechnol. 2022, 38, 252. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Jiang, W.-X.; Zhang, Y.-S.; Cao, H.-Y.; Zhang, Y.; Chen, X.-L.; Li, C.-Y.; Wang, P.; Zhang, Y.-Z.; Song, X.-Y.; et al. Structural Insight Into Chitin Degradation and Thermostability of a Novel Endochitinase from the Glycoside Hydrolase Family 18. Front. Microbiol. 2019, 10, 102457. [Google Scholar] [CrossRef]
- Vaghela, B.; Vashi, R.; Rajput, K.; Joshi, R. Plant chitinases and their role in plant defense: A comprehensive review. Enzym. Microb. Technol. 2022, 159, 110055. [Google Scholar] [CrossRef] [PubMed]
- Vandepas, L.E.; Tassia, M.G.; Halanych, K.M.; Amemiya, C.T. Unexpected Distribution of Chitin and Chitin Synthase across Soft-Bodied Cnidarians. Biomolecules 2023, 13, 777. [Google Scholar] [CrossRef]
- Grover, A. Plant Chitinases: Genetic Diversity and Physiological Roles. Crit. Rev. Plant Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Bravo, J.M.; Campo, S.; Murillo, I.; Coca, M.; San Segundo, B. Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family ofmaize. Plant Mol. Biol. 2003, 52, 745–759. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, W.; Zhang, X.; Huang, J.; Wang, W.; Miao, M.; Hu, L.; Wan, C.; Yuan, Y.; Wu, B.; et al. Genome-Wide Identification and Expression Analysis of Chitinase-like Genes in Petunia axillaris. Plants 2022, 11, 1269. [Google Scholar] [CrossRef]
- Berglund, L.; Brunstedt, J.; Nielsen, K.K.; Chen, Z.; Mikkelsen, J.D.; Marcker, K.A. A proline-rich chitinase from Beta vulgaris. Plant Mol. Biol. 1995, 27, 211–216. [Google Scholar] [CrossRef]
- Naumann, T.A.; Price, N.P.J. Truncation of class IV chitinases from Arabidopsis by secreted fungal proteases. Mol. Plant Pathol. 2012, 13, 1135–1139. [Google Scholar] [CrossRef]
- Richa, K.; Tiwari, I.M.; Devanna, B.N.; Botella, J.R.; Sharma, V.; Sharma, T.R. Novel Chitinase Gene LOC_Os11g47510 from Indica Rice Tetep Provides Enhanced Resistance against Sheath Blight Pathogen Rhizoctonia solani in Rice. Front. Plant Sci. 2017, 8, 596. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Tan, X. Comprehensive Analysis of the Chitinase Family Genes in Tomato (Solanum lycopersicum). Plants 2019, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Cletus, J.; Balasubramanian, V.; Vashisht, D.; Sakthivel, N. Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol. Lett. 2013, 35, 1719–1732. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.S.; Iqbal, A.; Bibi, A.; Khalil, I.; Ul Islam, Z.; Jan, F.; Khalid, A.; Abdalla, A.N.; Wadood, A. Plant chitinases: Types, structural classification, antifungal potential and transgenic expression in plants for enhanced disease resistance. Plant Cell Tissue Organ Cult. 2024, 156, 100–115. [Google Scholar] [CrossRef]
- Ohno, T.; Armand, S.; Hata, T.; Nikaidou, N.; Henrissat, B.; Mitsutomi, M.; Watanabe, T. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J. Bacteriol. 1996, 178, 5065–5070. [Google Scholar] [CrossRef]
- Udaya Prakash, N.A.; Jayanthi, M.; Sabarinathan, R.; Kangueane, P.; Mathew, L.; Sekar, K. Evolution, Homology Conservation, and Identification of Unique Sequence Signatures in GH19 Family Chitinases. J. Mol. Evol. 2010, 70, 466–478. [Google Scholar] [CrossRef]
- Hong, D.E.; Yu, J.E.; Lee, J.W.; Son, D.J.; Lee, H.P.; Kim, Y.; Chang, J.Y.; Lee, D.W.; Lee, W.K.; Yun, J.; et al. A Natural CHI3L1—Targeting Compound, Ebractenoid F, Inhibits Lung Cancer Cell Growth and Migration and Induces Apoptosis by Blocking CHI3L1/AKT Signals. Molecules 2023, 28, 329. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Wu, P.-L. Gene Cloning, Characterization, and Molecular Simulations of a Novel Recombinant Chitinase from Chitinibacter Tainanensis CT01 Appropriate for Chitin Enzymatic Hydrolysis. Polymers 2020, 12, 1648. [Google Scholar] [CrossRef]
- Morimoto, Y.; Takahashi, S.; Isoda, Y.; Nokami, T.; Fukamizo, T.; Suginta, W.; Ohnuma, T. Kinetic and thermodynamic insights into the inhibitory mechanism of TMG-chitotriomycin on Vibrio campbellii GH20 exo-β-N-acetylglucosaminidase. Carbohydr. Res. 2021, 499, 108201. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, N.; Chen, X.; Wei, G.; Zhang, A.; Chen, K.; Ouyang, P. Characterization of a New Multifunctional GH20 β-N-Acetylglucosaminidase from Chitinibacter sp. GC72 and Its Application in Converting Chitin Into N-Acetyl Glucosamine. Front. Microbiol. 2022, 13, 874908. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Tyagi, B.P.C.; Chakraborty, O.; Kumar, A.; Jain, A.K. Structural and functional analysis of chitinase gene family in wheat (Triticum aestivum). Indian J. Biochem. Biophys. 2015, 4, 169–178. [Google Scholar]
- Passarinho, P.A.; de Vries, S.C. ArabidopsisChitinases: A Genomic Survey. Arab. Book 2002, 1, e0023. [Google Scholar] [CrossRef] [PubMed]
- Lv, P.; Zhang, C.; Xie, P.; Yang, X.; El-Sheikh, M.A.; Hefft, D.I.; Ahmad, P.; Zhao, T.; Bhat, J.A. Genome-Wide Identification and Expression Analyses of the Chitinase Gene Family in Response to White Mold and Drought Stress in Soybean (Glycine max). Life 2022, 12, 1340. [Google Scholar] [CrossRef] [PubMed]
- Krishnaveni, S.; Liang, G.H.; Muthukrishnan, S.; Manickam, A. Purification and partial characterization of chitinases from sorghum seeds. Plant Sci. 1999, 144, 1–7. [Google Scholar] [CrossRef]
- Li, X.-m.; Chen, X.; Zhao, D.-G. Overexpression of the Eucommia ulmoides chitinase EuCHIT73.88 gene improves tobacco disease resistance. Gene 2024, 927, 148619. [Google Scholar] [CrossRef]
- Su, Y.; Xu, L.; Wang, S.; Wang, Z.; Yang, Y.; Chen, Y.; Que, Y. Identification, Phylogeny and Transcript of Chitinase Family Genes in Sugarcane. Sci. Rep. 2015, 5, 10708. [Google Scholar] [CrossRef]
- Bartholomew, E.S.; Black, K.; Feng, Z.; Liu, W.; Shan, N.; Zhang, X.; Wu, L.; Bailey, L.; Zhu, N.; Qi, C.; et al. Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum. Int. J. Mol. Sci. 2019, 20, 5309. [Google Scholar] [CrossRef]
- Ali, M.; Luo, D.-X.; Khan, A.; Haq, S.u.; Gai, W.-X.; Zhang, H.-X.; Cheng, G.-X.; Muhammad, I.; Gong, Z.-H. Classification and Genome-Wide Analysis of Chitin-Binding Proteins Gene Family in Pepper (Capsicum annuum L.) and Transcriptional Regulation to Phytophthora capsici, Abiotic Stresses and Hormonal Applications. Int. J. Mol. Sci. 2018, 19, 2216. [Google Scholar] [CrossRef]
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Nazari, L.; Zinati, Z.; Doherty, C. Transcriptional survey of abiotic stress response in maize (Zea mays) in the level of gene co-expression network and differential gene correlation analysis. AoB Plants 2024, 16, plad087. [Google Scholar] [CrossRef] [PubMed]
- Zha, H.-G.; Milne, R.I.; Zhou, H.-X.; Chen, X.-Y.; Sun, H. Identification and cloning of class II and III chitinases from alkaline floral nectar of Rhododendron irroratum, Ericaceae. Planta 2016, 244, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Z.; Zhang, W.Y.; Lin, Y.Z.; Li, D.Q.; Shu, B.S.; Lin, J.T. Genome-wide identification, characterization and functional analysis of the chitianse and chitinase-like gene family in Diaphorina citri. Pest. Manag. Sci. 2022, 78, 1740–1748. [Google Scholar] [CrossRef] [PubMed]
- Dowd, P.F.; Naumann, T.A.; Price, N.P.J.; Johnson, E.T. Identification of a maize (Zea mays) chitinase allele sequence suitable for a role in ear rot fungal resistance. Agri. Gene 2018, 7, 15–22. [Google Scholar] [CrossRef]
- Nawaz, M.; Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A review of plants strategies to resist biotic and abiotic environmental stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef]
- López, R.C.; Gómez-Gómez, L. Isolation of a new fungi and wound-induced chitinase class in corms of Crocus sativus. Plant Physiol. Biochem. 2009, 47, 426–434. [Google Scholar] [CrossRef]
- Gao, Y.; Zan, X.-L.; Wu, X.-F.; Yao, L.; Chen, Y.-L.; Jia, S.-W.; Zhao, K.-J. Identification of fungus-responsive cis-acting element in the promoter of Brassica juncea chitinase gene, BjCHI1. Plant Sci. 2014, 215–216, 190–198. [Google Scholar] [CrossRef]
- Tapia, G.; Morales-Quintana, L.; Inostroza, L.; Acuña, H. Molecular characterisation of Ltchi7, a gene encoding a Class III endochitinase induced by drought stress in Lotus spp. Plant Biol. 2010, 13, 69–77. [Google Scholar] [CrossRef]
- Cao, S.; Wang, Y.; Li, Z.; Shi, W.; Gao, F.; Zhou, Y.; Zhang, G.; Feng, J. Genome-Wide Identification and Expression Analyses of the Chitinases under Cold and Osmotic Stress in Ammopiptanthus nanus. Genes 2019, 10, 472. [Google Scholar] [CrossRef]
- Regalado, A.P.; Pinheiro, C.; Vidal, S.; Chaves, I.; Ricardo, C.P.P.; Rodrigues-Pousada, C. The Lupinus albus class-III chitinase gene, IF3, is constitutively expressed in vegetative organs and developing seeds. Planta 2000, 210, 543–550. [Google Scholar] [CrossRef]
- Békésiová, B.; Hraška, Š.; Libantová, J.; Moravčíková, J.; Matušíková, I. Heavy-metal stress induced accumulation of chitinase isoforms in plants. Mol. Biol. Rep. 2007, 35, 579–588. [Google Scholar] [CrossRef]
- Zhou, Y.-Y.; Wang, Y.-S.; Sun, C.-C.; Fei, J. Cloning and Expression of Class I Chitinase Genes from Four Mangrove Species under Heavy Metal Stress. Plants 2023, 12, 2772. [Google Scholar] [CrossRef]
- Abd El-Wahed, M.H.; Ali, E.A. Effect of irrigation systems, amounts of irrigation water and mulching on corn yield, water use efficiency and net profit. Agric. Water Manag. 2013, 120, 64–71. [Google Scholar] [CrossRef]
- Zhang, H. Effects of Soybean–Corn Rotation on Crop Yield, Economic Benefits, and Water Productivity in the Corn Belt of Northeast China. Sustainability 2023, 15, 1362. [Google Scholar] [CrossRef]
- Li, J.; Yang, S.; Yang, X.; Wu, H.; Tang, H.; Yang, L. PlantGF: An analysis and annotation platform for plant gene families. Database 2022, 2022, baab088. [Google Scholar] [CrossRef]
- Gabaldon, T.; Huynen, M.A. Prediction of protein function and pathways in the genome era. Cell. Mol. Life Sci. 2004, 61, 930–944. [Google Scholar] [CrossRef]
- Fan, M.; Gao, S.; Yang, Y.; Yang, S.; Wang, H.; Shi, L. Genome-wide identification and expression analysis of the universal stress protein (USP) gene family in Arabidopsis thaliana, Zea mays, and Oryza sativa. Genetica 2024, 152, 119–132. [Google Scholar] [CrossRef]
- Yan, Z.; Hou, J.; Leng, B.; Yao, G.; Ma, C.; Sun, Y.; Zhang, F.; Mu, C.; Liu, X. Genome-Wide Investigation of the CRF Gene Family in Maize and Functional Analysis of ZmCRF9 in Response to Multiple Abiotic Stresses. Int. J. Mol. Sci. 2024, 25, 7650. [Google Scholar] [CrossRef]
- Fang, H.; Shan, T.; Gu, H.; Chen, J.; Qi, Y.; Li, Y.; Saeed, M.; Yuan, J.; Li, P.; Wang, B. Identification and characterization of ACR gene family in maize for salt stress tolerance. Front. Plant Sci. 2024, 15, 11–20. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Y.; Zou, K.; Guan, M.; Wu, Y.; Hu, Y.; Yu, H.; Du, J.; Wu, D. The Analysis, Description, and Examination of the Maize LAC Gene Family’s Reaction to Abiotic and Biotic Stress. Genes 2024, 15, 749. [Google Scholar] [CrossRef]
- Song, Z.; Li, S.; Li, Y.; Zhou, X.; Liu, X.; Yang, W.; Chen, R. Identification and characterization of yellow stripe-like genes in maize suggest their roles in the uptake and transport of zinc and iron. BMC Plant Biol. 2024, 24, 3. [Google Scholar] [CrossRef]
- Shoresh, M.; Harman, G.E. Genome-wide identification, expression and chromosomal location of the genes encoding chitinolytic enzymes in Zea mays. Mol. Genet. Genom. 2008, 280, 173–185. [Google Scholar] [CrossRef]
- Coulombe, R.A.; Hawkins, L.K.; Mylroie, J.E.; Oliveira, D.A.; Smith, J.S.; Ozkan, S.; Windham, G.L.; Williams, W.P.; Warburton, M.L. Characterization of the Maize Chitinase Genes and Their Effect on Aspergillus flavus and Aflatoxin Accumulation Resistance. PLoS ONE 2015, 10, e0126185. [Google Scholar] [CrossRef]
- Cazares-Álvarez, J.E.; Báez-Astorga, P.A.; Arroyo-Becerra, A.; Maldonado-Mendoza, I.E. Genome-Wide Identification of a Maize Chitinase Gene Family and the Induction of Its Expression by Fusarium verticillioides (Sacc.) Nirenberg (1976) Infection. Genes 2024, 15, 1087. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2007, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2011, 40, D290–D301. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Brown, G.R.; Maglott, D.R. NCBI Reference Sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Res. 2011, 40, D130–D135. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Pan, X.; Hu, M.; Wang, Z.; Su, M.; Lei, K.; Wu, H.; Jiang, X. Genome-wide identification and expression analysis of rice chitinase gene family. J. Plant Physiol. 2022, 58, 746–756. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Poon, A.F.Y.; Zhang, Y.-J.; Ma, P.-F.; Li, D.-Z. High-Throughput Sequencing of Six Bamboo Chloroplast Genomes: Phylogenetic Implications for Temperate Woody Bamboos (Poaceae: Bambusoideae). PLoS ONE 2011, 6, e20596. [Google Scholar] [CrossRef]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Feng, Q.; Gao, Z.; Lu, H.; Hu, T.; Yao, N.; Liu, K.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R.; Valencia, A. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Yin, Y.; Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A.; Wren, J. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Sun, M.-x.; Sekhon, R.S.; Briskine, R.; Hirsch, C.N.; Myers, C.L.; Springer, N.M.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Maize Gene Atlas Developed by RNA Sequencing and Comparative Evaluation of Transcriptomes Based on RNA Sequencing and Microarrays. PLoS ONE 2013, 8, e61005. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Li, Y.; Zhang, Y.; Gou, Z.; Qi, X.; Zhang, J. Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses. Genes 2020, 11, 881. [Google Scholar] [CrossRef]
- Yu, F.; Tan, Z.; Fang, T.; Tang, K.; Liang, K.; Qiu, F. A Comprehensive Transcriptomics Analysis Reveals Long Non-Coding RNA to Be Involved in the Key Metabolic Pathway in Response to Waterlogging Stress in Maize. Genes 2020, 11, 267. [Google Scholar] [CrossRef]
- Prasad, M.; Jin, H.; Liu, S.; Zenda, T.; Wang, X.; Liu, G.; Duan, H. Maize leaves drought-responsive genes revealed by comparative transcriptome of two cultivars during the filling stage. PLoS ONE 2019, 14, e0223786. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Zhang, Y.; Li, C.; Gong, S.; Yan, S.; Li, G.; Hu, G.; Ren, H.; Yang, J.; et al. Comparative transcriptome analysis of salt-sensitive and salt-tolerant maize reveals potential mechanisms to enhance salt resistance. Genes Genom. 2019, 41, 781–801. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Gou, X.; Zhang, W.; Li, T.; Xue, J.; Guo, D.; Xu, S. New insights into the response of maize to fluctuations in the light environment. Mol. Genet. Genom. 2021, 296, 615–629. [Google Scholar] [CrossRef]
- Schurack, S.; Depotter, J.R.L.; Gupta, D.; Thines, M.; Doehlemann, G. Comparative transcriptome profiling identifies maize line specificity of fungal effectors in the maize–Ustilago maydis interaction. Plant J. 2021, 106, 733–752. [Google Scholar] [CrossRef]
- Sun, Y.; Ruan, X.; Wang, Q.; Zhou, Y.; Wang, F.; Ma, L.; Wang, Z.; Gao, X. Integrated Gene Co-expression Analysis and Metabolites Profiling Highlight the Important Role of ZmHIR3 in Maize Resistance to Gibberella Stalk Rot. Front. Plant Sci. 2021, 12, 664733. [Google Scholar] [CrossRef]
- Yu, Y.; Shi, J.; Li, X.; Liu, J.; Geng, Q.; Shi, H.; Ke, Y.; Sun, Q. Transcriptome analysis reveals the molecular mechanisms of the defense response to gray leaf spot disease in maize. BMC Genom. 2018, 19, 742. [Google Scholar] [CrossRef]
- Tzin, V.; Hojo, Y.; Strickler, S.R.; Bartsch, L.J.; Archer, C.M.; Ahern, K.R.; Zhou, S.; Christensen, S.A.; Galis, I.; Mueller, L.A.; et al. Rapid defense responses in maize leaves induced by Spodoptera exigua caterpillar feeding. J. Exp. Bot. 2017, 68, 4709–4723. [Google Scholar] [CrossRef]
- Tang, Y.; Guo, J.; Zhang, T.; Bai, S.; He, K.; Wang, Z. Genome-Wide Analysis of WRKY Gene Family and the Dynamic Responses of Key WRKY Genes Involved in Ostrinia furnacalis Attack in Zea mays. Int. J. Mol. Sci. 2021, 22, 13045. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Fernandez-Pozo, N.; Richter, A.; Schmelz, E.A.; Schoettner, M.; Schäfer, M.; Ahern, K.R.; Meihls, L.N.; Kaur, H.; Huffaker, A.; et al. Dynamic maize responses to aphid feeding are revealed by a time series of transcriptomic and metabolomic assays. Plant Physiol. 2015, 169, 1727–1743. [Google Scholar] [CrossRef]
- Zhang, H.; Wafula, E.K.; Eilers, J.; Harkess, A.E.; Ralph, P.E.; Timilsena, P.R.; dePamphilis, C.W.; Waite, J.M.; Honaas, L.A. Building a foundation for gene family analysis in Rosaceae genomes with a novel workflow: A case study in Pyrus architecture genes. Front. Plant Sci. 2022, 13, 975942. [Google Scholar] [CrossRef]
- Lv, Z.; Jiang, S.; Kong, S.; Zhang, X.; Yue, J.; Zhao, W.; Li, L.; Lin, S. Advances in Single-Cell Transcriptome Sequencing and Spatial Transcriptome Sequencing in Plants. Plants 2024, 13, 1679. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, M.; Wei, X.; Xia, R.; Pei, D.; Huang, X.; Han, B. Computational tools for plant genomics and breeding. Sci. China Life Sci. 2024, 67, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Cui, X.; Yu, X.; Ning, X.; Yu, H.; Li, J.; Yang, B.; Pan, Y.; Jiang, L. Molecular evolution and functional diversification of metal tolerance protein families in cereals plants and function of maize MTP protein. Int. J. Biol. Macromol. 2024, 274, 133071. [Google Scholar] [CrossRef] [PubMed]
- Hoa, P.T.B.; Phuong, H.L.; Loc, N.H. Characteristics on growth, development, and inheritance of 42 kDa chitinase-transgenic peanut lines in the T1 progeny. J. Crop Sci. Biotechnol. 2024, 14, 337–350. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Y.; Zou, H.; Li, X.; Zou, H.; Wang, Z.; Zou, C. FDP-Na-induced enhancement of glycolysis impacts larval growth and development and chitin biosynthesis in fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Pestic. Biochem. Physiol. 2023, 195, 105560. [Google Scholar] [CrossRef]
- Pan, L.; Wen, S.; Yu, J.; Lu, L.; Zhu, X.; Zhang, Z. Genome-Wide Identification of M35 Family Metalloproteases in Rhizoctonia cerealis and Functional Analysis of RcMEP2 as a Virulence Factor during the Fungal Infection to Wheat. Int. J. Mol. Sci. 2020, 21, 2984. [Google Scholar] [CrossRef]
- Davis, J.M.; Wu, H.; Cooke, J.E.K.; Reed, J.M.; Luce, K.S.; Michler, C.H. Pathogen Challenge, Salicylic Acid, and Jasmonic Acid Regulate Expression of Chitinase Gene Homologs in Pine. Mol. Plant-Microbe Interact. 2002, 15, 380–387. [Google Scholar] [CrossRef]
- Liu, X.; Yu, Y.; Liu, Q.; Deng, S.; Jin, X.; Yin, Y.; Guo, J.; Li, N.; Liu, Y.; Han, S.; et al. A Na2CO3-Responsive Chitinase Gene from Leymus chinensis Improve Pathogen Resistance and Saline-Alkali Stress Tolerance in Transgenic Tobacco and Maize. Front. Plant Sci. 2020, 11, 504. [Google Scholar] [CrossRef]
- Chu, F.; Wang, D.; Liu, T.; Han, H.; Yu, Y.; Yang, Q. An optimized cocktail of chitinolytic enzymes to produce N,N′-diacetylchitobiose and N-acetyl-d-glucosamine from defatted krill by-products. Int. J. Biol. Macromol. 2019, 133, 1029–1034. [Google Scholar] [CrossRef]
- Navarro-González, S.S.; Ramírez-Trujillo, J.A.; Peña-Chora, G.; Gaytán, P.; Roldán-Salgado, A.; Corzo, G.; Lina-García, L.P.; Hernández-Velázquez, V.M.; Suárez-Rodríguez, R. Enhanced Tolerance against a Fungal Pathogen and Insect Resistance in Transgenic Tobacco Plants Overexpressing an Endochitinase Gene from Serratia marcescens. Int. J. Mol. Sci. 2019, 20, 3482. [Google Scholar] [CrossRef]
- Sun, X.; Huo, L.; Jia, X.; Che, R.; Gong, X.; Wang, P.; Ma, F. Overexpression of MdATG18a in apple improves resistance to Diplocarpon mali infection by enhancing antioxidant activity and salicylic acid levels. Hortic. Res. 2018, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Parvin, W.; Govender, N.; Othman, R.; Jaafar, H.; Rahman, M.; Wong, M.-Y. Phenazine from Pseudomonas aeruginosa UPMP3 induced the host resistance in oil palm (Elaeis guineensis Jacq.)-Ganoderma boninense pathosystem. Sci. Rep. 2020, 10, 15621. [Google Scholar] [CrossRef] [PubMed]
- Passarinho, P.A.; Van Hengel, A.J.; Fransz, P.F.; de Vries, S.C. Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta 2001, 212, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, E.M.; Bovy, A.; Manning, K.; Harrison, L.; Andrews, J.; De Silva, J.; Tucker, G.A.; Seymour, G.B. Effect of the Colorless non-ripening Mutation on Cell Wall Biochemistry and Gene Expression during Tomato Fruit Development and Ripening. Plant Physiol. 2004, 136, 4184–4197. [Google Scholar] [CrossRef]
- Agustí, J.; Merelo, P.; Cercós, M.; Tadeo, F.R.; Talón, M. Ethylene-induced differential gene expression during abscission of citrus leaves. J. Exp. Bot. 2008, 59, 2717–2733. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Wang, C.; Liu, Y.; Zou, K.; Guan, M.; Wu, Y.; Yue, S.; Hu, Y.; Yu, H.; Zhang, K.; et al. Genome-Wide Identification of the Maize Chitinase Gene Family and Analysis of Its Response to Biotic and Abiotic Stresses. Genes 2024, 15, 1327. https://doi.org/10.3390/genes15101327
Wang T, Wang C, Liu Y, Zou K, Guan M, Wu Y, Yue S, Hu Y, Yu H, Zhang K, et al. Genome-Wide Identification of the Maize Chitinase Gene Family and Analysis of Its Response to Biotic and Abiotic Stresses. Genes. 2024; 15(10):1327. https://doi.org/10.3390/genes15101327
Chicago/Turabian StyleWang, Tonghan, Changjin Wang, Yang Liu, Kunliang Zou, Minghui Guan, Yutong Wu, Shutong Yue, Ying Hu, Haibing Yu, Kaijing Zhang, and et al. 2024. "Genome-Wide Identification of the Maize Chitinase Gene Family and Analysis of Its Response to Biotic and Abiotic Stresses" Genes 15, no. 10: 1327. https://doi.org/10.3390/genes15101327
APA StyleWang, T., Wang, C., Liu, Y., Zou, K., Guan, M., Wu, Y., Yue, S., Hu, Y., Yu, H., Zhang, K., Wu, D., & Du, J. (2024). Genome-Wide Identification of the Maize Chitinase Gene Family and Analysis of Its Response to Biotic and Abiotic Stresses. Genes, 15(10), 1327. https://doi.org/10.3390/genes15101327





