A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs
2.2. Cell Culture, Plasmid, and siRNA Transfections
2.3. Isolation of Total RNA and Protein Lysates
2.4. Western Blot Analysis
2.5. Reverse Transcription, Semi-Quantitative PCR (sqPCR), and Real-Time PCR (RT-qPCR)
2.6. Statistical Analysis
3. Results
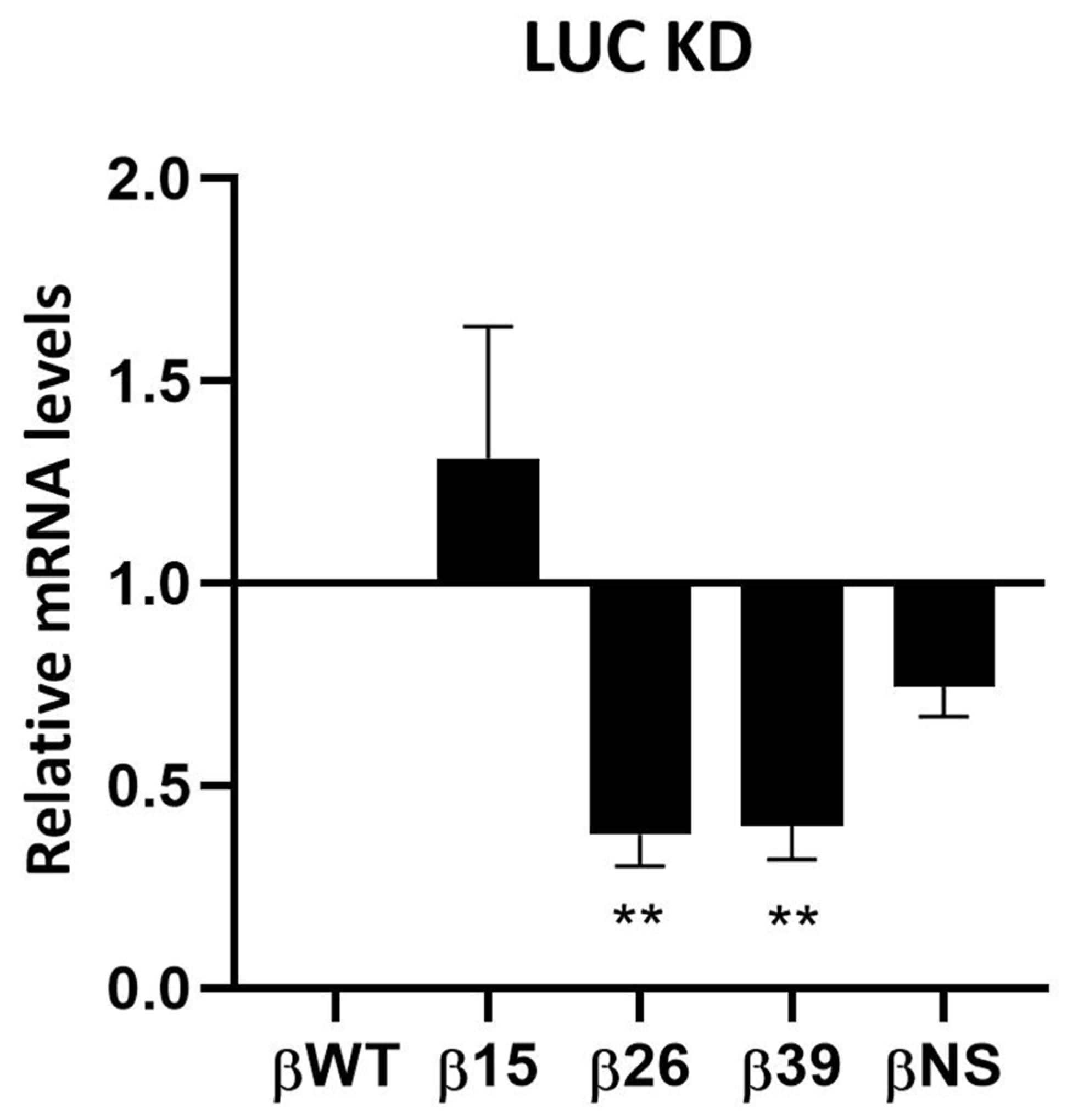
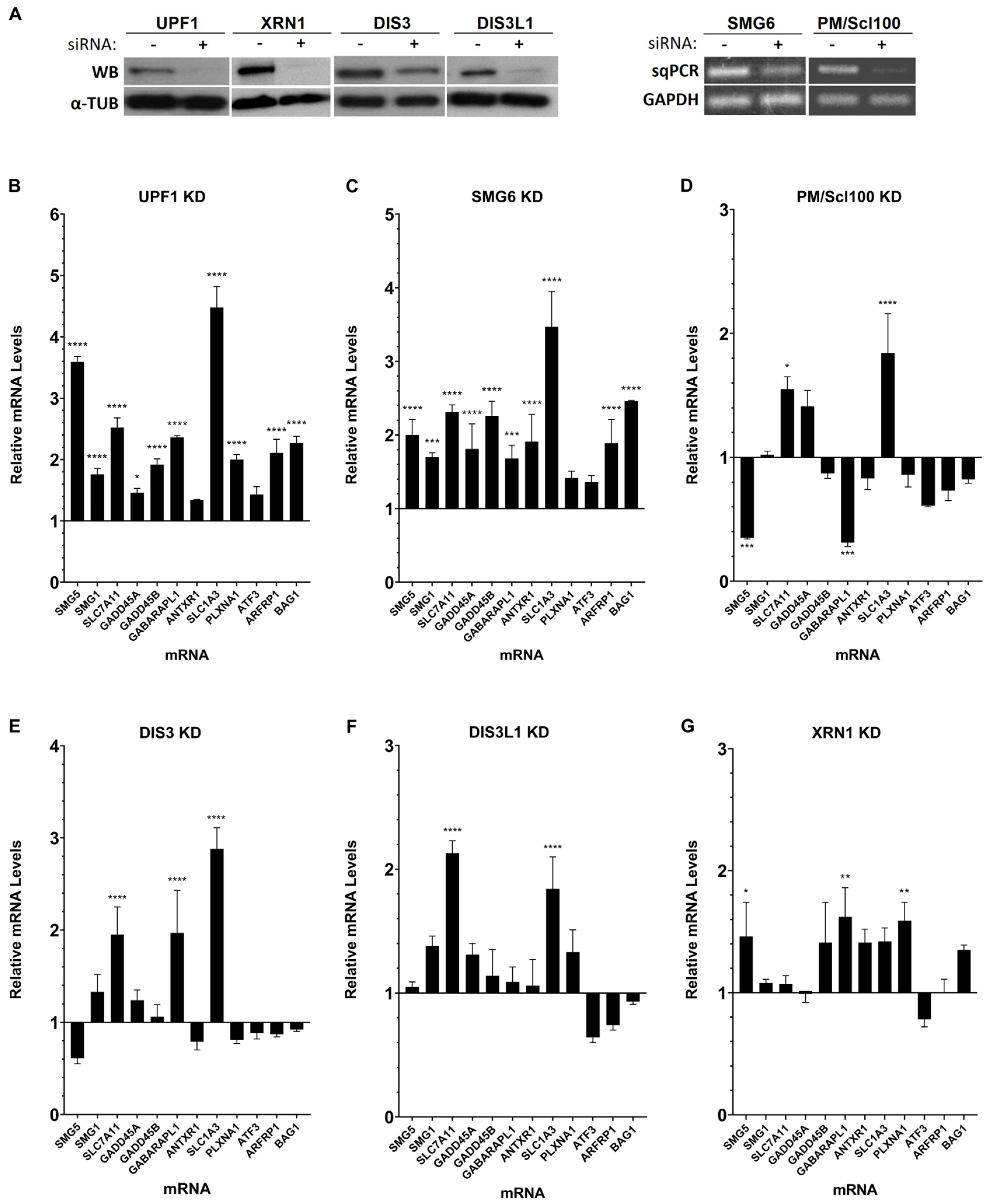
3.1. Human Ribonucleases Modulate the mRNA Levels of Different Natural NMD Targets
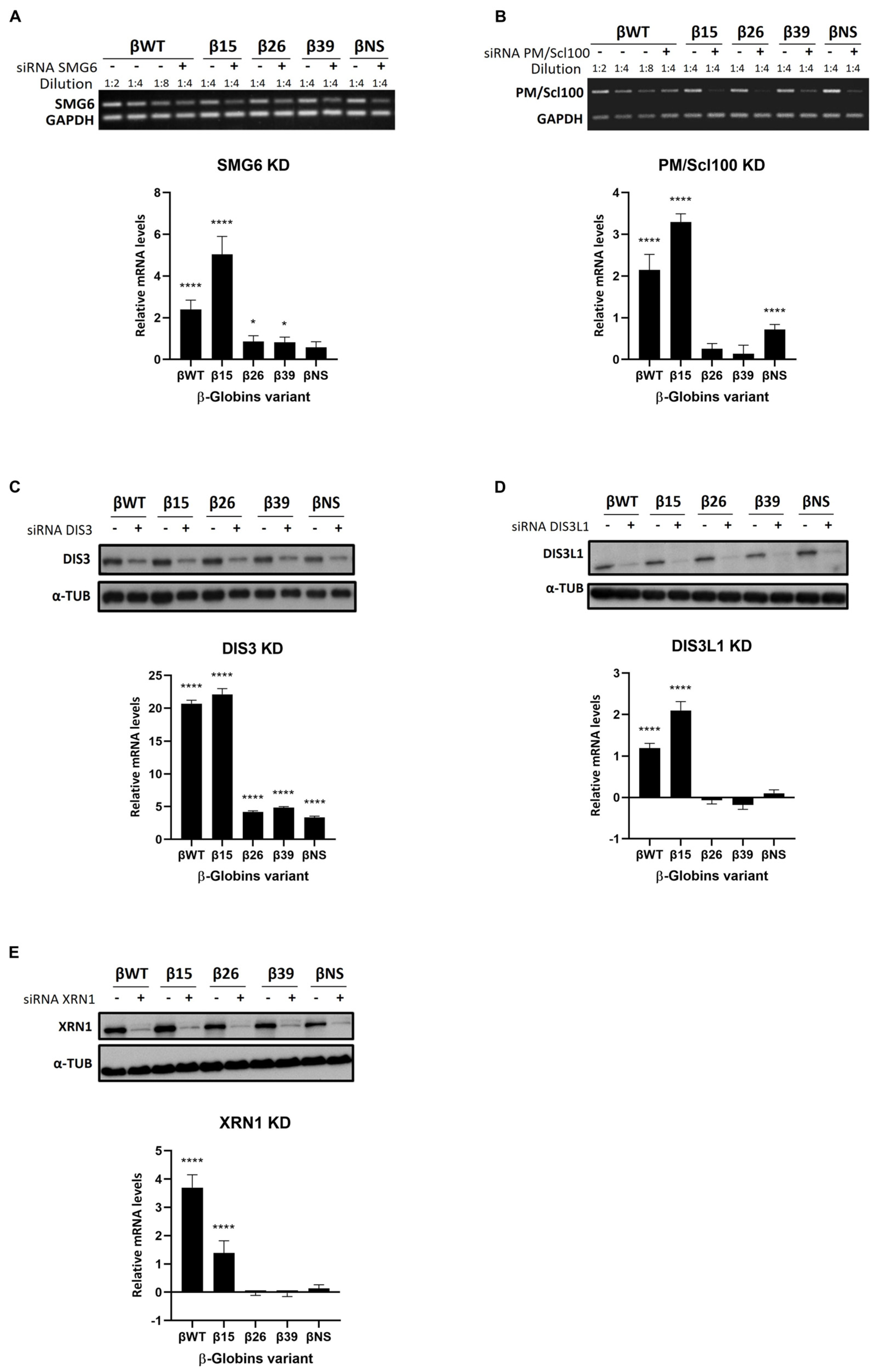
3.2. Roles of Human Ribonucleases in mRNA Surveillance Mechanisms: Use of Reporter Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Linde, L.; Kerem, B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008, 24, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Bhuvanagiri, M.; Schlitter, A.M.; Hentze, M.W.; Kulozik, A.E. NMD: RNA biology meets human genetic medicine. Biochem. J. 2010, 430, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Fasken, M.B.; Corbett, A.H. Process or perish: Quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 2005, 12, 482–488. [Google Scholar] [CrossRef]
- Klauer, A.A.; van Hoof, A. Degradation of mRNAs that lack a stop codon: A decade of nonstop progress. Wiley Interdiscip. Rev. RNA 2012, 3, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Schweingruber, C.; Rufener, S.C.; Zünd, D.; Yamashita, A.; Mühlemann, O. Nonsense-mediated mRNA decay—Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta-Gene Regul. Mech. 2013, 1829, 612–623. [Google Scholar] [CrossRef]
- Silva, A.L.; Romão, L. The mammalian nonsense-mediated mRNA decay pathway: To decay or not to decay! Which players make the decision? FEBS Lett. 2009, 583, 499–505. [Google Scholar] [CrossRef]
- Mühlemann, O.; Jensen, T.H. mRNP quality control goes regulatory. Trends Genet. 2012, 28, 70–77. [Google Scholar] [CrossRef]
- Saito, S.; Hosoda, N.; Hoshino, S.-I. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem. 2013, 288, 17832–17843. [Google Scholar] [CrossRef]
- Mendell, J.T.; A Sharifi, N.; Meyers, J.L.; Martinez-Murillo, F.; Dietz, H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004, 36, 1073–1078. [Google Scholar] [CrossRef]
- Nicholson, P.; Yepiskoposyan, H.; Metze, S.; Orozco, R.Z.; Kleinschmidt, N.; Mühlemann, O. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 2009, 67, 677–700. [Google Scholar] [CrossRef]
- Mühlemann, O.; Lykke-Andersen, J. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol. 2010, 7, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Kashima, I.; Fauser, M.; Saulière, J.; Izaurralde, E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 2008, 14, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Eberle, A.B.; Lykke-Andersen, S.; Mühlemann, O.; Jensen, T.H. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009, 16, 49–55. [Google Scholar] [CrossRef]
- Lejeune, F.; Li, X.; Maquat, L.E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003, 12, 675–687. [Google Scholar] [CrossRef]
- Chen, C.-Y.A.; Shyu, A.-B. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell Biol. 2003, 23, 4805–4813. [Google Scholar] [CrossRef]
- Couttet, P.; Grange, T. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res. 2004, 32, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Jonas, S.; Izaurralde, E. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013, 27, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Brodersen, D.E.; Jensen, T.H. Origins and activities of the eukaryotic exosome. J. Cell Sci. 2009, 122, 1487–1494. [Google Scholar] [CrossRef][Green Version]
- Mitchell, P.; Petfalski, E.; Shevchenko, A.; Mann, M.; Tollervey, D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 1997, 91, 457–466. [Google Scholar] [CrossRef]
- Allmang, C.; Petfalski, E.; Podtelejnikov, A.; Mann, M.; Tollervey, D.; Mitchell, P. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 1999, 13, 2148–2158. [Google Scholar] [CrossRef]
- Liu, Q.; Greimann, J.C.; Lima, C.D. Reconstitution, Activities, and Structure of the Eukaryotic RNA Exosome. Cell 2006, 127, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Dziembowski, A.; Lorentzen, E.; Conti, E.; Séraphin, B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007, 14, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Frazão, C.; McVey, C.E.; Amblar, M.; Barbas, A.; Vonrhein, C.; Arraiano, C.M.; Carrondo, M.A. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 2006, 443, 110–114. [Google Scholar] [CrossRef]
- Zuo, Y.; Vincent, H.A.; Zhang, J.; Wang, Y.; Deutscher, M.P.; Malhotra, A. Structural Basis for Processivity and Single-Strand Specificity of RNase II. Mol. Cell 2006, 24, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, A.; Tomecki, R.; Dziembowski, A.; Séraphin, B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 2008, 456, 993–996. [Google Scholar] [CrossRef]
- Schaeffer, D.; Tsanova, B.; Barbas, A.; Reis, F.P.; Dastidar, E.G.; Sanchez-Rotunno, M.; Arraiano, C.M.; van Hoof, A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009, 16, 56–62. [Google Scholar] [CrossRef]
- Staals, R.H.J.; Bronkhorst, A.W.; Schilders, G.; Slomovic, S.; Schuster, G.; Heck, A.J.R.; Raijmakers, R.; Pruijn, G.J.M. Dis3-like 1, A novel exoribonuclease associated with the human exosome. EMBO J. 2010, 29, 2358–2367. [Google Scholar] [CrossRef]
- Tomecki, R.; Kristiansen, M.S.; Lykke-Andersen, S.; Chlebowski, A.; Larsen, K.M.; Szczesny, R.J.; Drazkowska, K.; Pastula, A.; Andersen, J.S.; Stepien, P.P.; et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010, 29, 2342–2357. [Google Scholar] [CrossRef]
- Ustianenko, D.; Hrossova, D.; Potesil, D.; Chalupnikova, K.; Hrazdilova, K.; Pachernik, J.; Cetkovska, K.; Uldrijan, S.; Zdrahal, Z.; Vanacova, S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 2013, 19, 1632–1638. [Google Scholar] [CrossRef]
- Malecki, M.; Viegas, S.C.; Carneiro, T.; Golik, P.; Dressaire, C.; Ferreira, M.G.; Arraiano, C.M. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013, 32, 1842–1854. [Google Scholar] [CrossRef]
- Chang, H.-M.; Triboulet, R.; Thornton, J.E.; Gregory, R.I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013, 497, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Ha, M.; Chang, H.; Kwon, S.C.; Simanshu, D.K.; Patel, D.J.; Kim, V.N. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 2014, 159, 1365–1376. [Google Scholar] [CrossRef]
- Thomas, M.P.; Liu, X.; Whangbo, J.; McCrossan, G.; Sanborn, K.B.; Basar, E.; Walch, M.; Lieberman, J. Apoptosis Triggers Specific, Rapid, and Global mRNA Decay with 3′ Uridylated Intermediates Degraded by DIS3L2. Cell Rep. 2015, 11, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Ustianenko, D.; Pasulka, J.; Feketova, Z.; Bednarik, L.; Zigackova, D.; Fortova, A.; Zavolan, M.; Vanacova, S. TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J. 2016, 35, 2179–2191. [Google Scholar] [CrossRef]
- da Costa, P.J.; Menezes, J.; Saramago, M.; García-Moreno, J.F.; Santos, H.A.; Gama-Carvalho, M.; Arraiano, C.M.; Viegas, S.C.; Romão, L. A role for DIS3L2 over natural nonsense-mediated mRNA decay targets in human cells. Biochem. Biophys. Res. Commun. 2019, 518, 664–671. [Google Scholar] [CrossRef]
- Kurosaki, T.; Miyoshi, K.; Myers, J.R.; Maquat, L.E. NMD-degradome sequencing reveals ribosome-bound intermediates with 3′-end non-templated nucleotides. Nat. Struct. Mol. Biol. 2018, 25, 940. [Google Scholar] [CrossRef]
- Romão, L.; Inácio, Â.; Santos, S.; Ávila, M.; Faustino, P.; Pacheco, P.; Lavinha, J. Nonsense mutations in the human β-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation. Blood 2000, 96, 2895–2902. [Google Scholar] [CrossRef] [PubMed]
- Inácio, Â.; Silva, A.L.; Pinto, J.; Ji, X.; Morgado, A.; Almeida, F.; Faustino, P.; Lavinha, J.; Liebhaber, S.A.; Romão, L. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J. Biol. Chem. 2004, 279, 32170–32180. [Google Scholar] [CrossRef]
- Silva, A.L.; Ribeiro, P.; Inácio, Â.; Liebhaber, S.A.; Romão, L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 2008, 14, 563–576. [Google Scholar] [CrossRef]
- da Costa, P.J.; Menezes, J.; Saramago, M.; García-Moreno, J.F.; Santos, H.A.; Gama-Carvalho, M.; Arraiano, C.M.; Viegas, S.C.; Romão, L. A role for DIS3L2 over human nonsense-mediated mRNA decay targets. bioRxiv 2019. [Google Scholar] [CrossRef]
- Brogna, S.; McLeod, T.; Petric, M. The Meaning of NMD: Translate or Perish. Trends Genet. 2016, 32, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Hug, N.; Cáceres, J.F. The RNA Helicase DHX34 Activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep. 2014, 8, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Karousis, E.D.; Nasif, S.; Mühlemann, O. Nonsense-mediated mRNA decay: Novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA 2016, 7, 661–682. [Google Scholar] [CrossRef]
- Saveanu, C.; Jacquier, A. How cells kill a “killer” messenger. eLife 2016, 5, e16076. [Google Scholar] [CrossRef]
- Celik, A.; Baker, R.; He, F.; Jacobson, A. High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA 2017, 23, 735–748. [Google Scholar] [CrossRef]
- Karousis, E.D.; Mühlemann, O. Nonsense-Mediated mRNA Decay Begins where Translation Ends. Cold Spring Harb. Perspect. Biol. 2019, 11, a032862. [Google Scholar] [CrossRef] [PubMed]
- Raimondeau, E.; Bufton, J.C.; Schaffitzel, C. New insights into the interplay between the translation machinery and nonsense-mediated mRNA decay factors. Biochem. Soc. Trans. 2018, 46, 503–512. [Google Scholar] [CrossRef]
- Kim, Y.K.I.; Maquat, L.E. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 2019, 25, 407–422. [Google Scholar] [CrossRef]
- Tani, H.; Imamachi, N.; Salam, K.A.; Mizutani, R.; Ijiri, K.; Irie, T.; Yada, T.; Suzuki, Y.; Akimitsu, N. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 2012, 9, 1370–1379. [Google Scholar] [CrossRef]
- Boehm, V.; Kueckelmann, S.; Gerbracht, J.V.; Kallabis, S.; Britto-Borges, T.; Altmüller, J.; Krüger, M.; Dieterich, C.; Gehring, N.H. SMG5-SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity. Nat. Commun. 2021, 12, 1–19. [Google Scholar] [CrossRef]
- Yamashita, A. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells 2013, 18, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Nath, P.; Alfarsi, L.H.; El-Ansari, R.; Masisi, B.K.; Erkan, B.; Fakroun, A.; Ellis, I.O.; Rakha, E.A.; Green, A.R. The amino acid transporter SLC7A11 expression in breast cancer. Cancer Biol. Ther. 2024, 25, 2291855. [Google Scholar] [CrossRef]
- Andersen, J.V.; Markussen, K.H.; Jakobsen, E.; Schousboe, A.; Waagepetersen, H.S.; Rosenberg, P.A.; Aldana, B.I. Glutamate metabolism and recycling at the excitatory synapse in health and neurodegeneration. Neuropharmacology 2021, 196, 108719. [Google Scholar] [CrossRef]
- Tamura, R.E.; De Vasconcellos, J.F.; Sarkar, D.; Libermann, T.A.; Fisher, P.B.; Zerbini, L.F. GADD45 proteins: Central players in tumorigenesis. Curr. Mol. Med. 2012, 12, 634–651. [Google Scholar] [CrossRef]
- Hervouet, E.; Claude-Taupin, A.; Gauthier, T.; Perez, V.; Fraichard, A.; Adami, P.; Despouy, G.; Monnien, F.; Algros, M.-P.; Jouvenot., M.; et al. The autophagy GABARAPL1 gene is epigenetically regulated in breast cancer models. BMC Cancer 2015, 15, 729. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, J.; Zheng, Y. ANTXR1 Is a Prognostic Biomarker and Correlates with Stromal and Immune Cell Infiltration in Gastric Cancer. Front. Mol. Biosci. 2020, 7, 598221. [Google Scholar] [CrossRef]
- Hai, T.; Wolfgang, C.D.; Marsee, D.K.; Allen, A.E.; Sivaprasad, U. ATF3 and Stress Responses. Gene Expr. 1999, 7, 321. [Google Scholar]
- Hommel, A.; Hesse, D.; Völker, W.; Jaschke, A.; Moser, M.; Engel, T.; Blüher, M.; Zahn, C.; Chadt, A.; Ruschke, K.; et al. The ARF-like GTPase ARFRP1 is essential for lipid droplet growth and is involved in the regulation of lipolysis. Mol. Cell Biol. 2010, 30, 1231–1242. [Google Scholar] [CrossRef]
- Cutress, R.I.; Townsend, P.A.; Brimmell, M.; Bateman, A.C.; Hague, A.; Packham, G. BAG-1 expression and function in human cancer. Br. J. Cancer 2002, 87, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Boehm, V.; Haberman, N.; Ottens, F.; Ule, J.; Gehring, N.H. 3′ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep. 2014, 9, 555–568. [Google Scholar] [CrossRef]
- Schmidt, S.A.; Foley, P.L.; Jeong, D.-H.; Rymarquis, L.A.; Doyle, F.; Tenenbaum, S.A.; Belasco, J.G.; Green, P.J. Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res. 2015, 43, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Karousis, E.D.; Bourquin, J.; Bruggmann, R.; Mühlemann, O. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 2017, 23, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Ottens, F.; Boehm, V.; Sibley, C.R.; Ule, J.; Gehring, N.H. Transcript-specific characteristics determine the contribution of endo- and exonucleolytic decay pathways during the degradation of nonsense-mediated decay substrates. RNA 2017, 23, 1224–1236. [Google Scholar] [CrossRef]
- Metze, S.; Herzog, V.A.; Ruepp, M.-D.; Mühlemann, O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 2013, 19, 1432–1448. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, P.; Josi, C.; Kurosawa, H.; Yamashita, A.; Mühlemann, O. A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res. 2014, 42, 9217–9235. [Google Scholar] [CrossRef]
- Gatfield, D.; Izaurralde, E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 2004, 429, 575–578. [Google Scholar] [CrossRef]
- Schaeffer, D.; van Hoof, A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 2366–2371. [Google Scholar] [CrossRef]
- Jamar, N.H.; Kritsiligkou, P.; Grant, C.M. The non-stop decay mRNA surveillance pathway is required for oxidative stress tolerance. Nucleic Acids Res. 2017, 45, 6881–6893. [Google Scholar] [CrossRef]
- Robinson, S.R.; Oliver, A.W.; Chevassut, T.J.; Newbury, S.F. The 3′ to 5′ Exoribonuclease DIS3, From Structure and Mechanisms to Biological Functions and Role in Human Disease. Biomolecules 2015, 5, 1515–1539. [Google Scholar] [CrossRef]
- Szczepińska, T.; Kalisiak, K.; Tomecki, R.; Labno, A.; Borowski, L.S.; Kulinski, T.M.; Adamska, D.; Kosinska, J.; Dziembowski, A. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015, 25, 1622–1633. [Google Scholar] [CrossRef]
- Andrulis, E.D.; Werner, J.; Nazarian, A.; Erdjument-Bromage, H.; Tempst, P.; Lis, J.T. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 2002, 420, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Hessle, V.; Björk, P.; Sokolowski, M.; de Valdivia, E.G.; Silverstein, R.; Artemenko, K.; Tyagi, A.; Maddalo, G.; Ilag, L.; Helbig, R.; et al. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol. Biol. Cell 2009, 20, 3459–3470. [Google Scholar] [CrossRef] [PubMed]
- van Hoof, A.; Lennertz, P.; Parker, R. Yeast Exosome Mutants Accumulate 3′-Extended Polyadenylated Forms of U4 Small Nuclear RNA and Small Nucleolar RNAs. Mol. Cell Biol. 2000, 20, 441–452. [Google Scholar] [CrossRef]
- Januszyk, K.; Lima, C.D. The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 2014, 24, 132–140. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, E.L.; Schilders, G.; Pruijn, G.J.M. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA 2007, 13, 1027–1035. [Google Scholar] [CrossRef]
- Wasmuth, E.V.; Lima, C.D. The Rrp6 C-terminal domain binds RNA and activates the nuclear RNA exosome. Nucleic Acids Res. 2017, 45, 846–860. [Google Scholar] [CrossRef]
- Wasmuth, E.V.; Lima, C.D. Structure and Activities of the Eukaryotic RNA Exosome. In The Enzymes; Academic Press: Cambridge, MA, USA, 2012; pp. 53–75. [Google Scholar] [CrossRef]
- Wasmuth, E.V.; Lima, C.D. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell 2012, 48, 133–144. [Google Scholar] [CrossRef]
- Haimovich, G.; Medina, D.A.; Causse, S.Z.; Garber, M.; Millán-Zambrano, G.; Barkai, O.; Chávez, S.; Pérez-Ortín, J.E.; Darzacq, X.; Choder, M. Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell 2013, 153, 1000–1011. [Google Scholar] [CrossRef]
- Sun, M.; Schwalb, B.; Pirkl, N.; Maier, K.C.; Schenk, A.; Failmezger, H.; Tresch, A.; Cramer, P. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell 2013, 52, 52–62. [Google Scholar] [CrossRef]
- Jones, C.I.; Zabolotskaya, M.V.; Newbury, S.F. The 5′ → 3′ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip. Rev. RNA 2012, 3, 455–468. [Google Scholar] [CrossRef]
- Łabno, A.; Tomecki, R.; Dziembowski, A. Cytoplasmic RNA decay pathways—Enzymes and mechanisms. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 3125–3147. [Google Scholar] [CrossRef]
- Moon, S.L.; Blackinton, J.G.; Anderson, J.R.; Dozier, M.K.; Dodd, B.J.T.; Keene, J.D.; Wilusz, C.J.; Bradrick, S.S.; Wilusz, J. XRN1 stalling in the 5′ UTR of Hepatitis C virus and Bovine Viral Diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog. 2015, 11, e1004708. [Google Scholar] [CrossRef] [PubMed]
- MacFadden, A.; O’donoghue, Z.; Silva, P.A.G.C.; Chapman, E.G.; Olsthoorn, R.C.; Sterken, M.G.; Pijlman, G.P.; Bredenbeek, P.J.; Kieft, J.S. Mechanism and structural diversity of exoribonuclease-resistant RNA structures in flaviviral RNAs. Nat. Commun. 2018, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Vaché, C.; Cubedo, N.; Mansard, L.; Sarniguet, J.; Baux, D.; Faugère, V.; Baudoin, C.; Moclyn, M.; Touraine, R.; Lina-Granade, G.; et al. Identification and in vivo functional investigation of a HOMER2 nonstop variant causing hearing loss. Eur. J. Hum. Genet. 2023, 31, 834–840. [Google Scholar] [CrossRef]
- Terrey, M.; I Adamson, S.; Chuang, J.H.; Ackerman, S.L.; Neurobiology, S.O.; States, U. Defects in translation-dependent quality control pathways lead to convergent molecular and neurodevelopmental pathology. eLife 2021, 10, e66904. [Google Scholar] [CrossRef]
| Ribonucleases | ||||||
|---|---|---|---|---|---|---|
| Targets | SMG6 a | PM/Scl100 a | DIS3 a | DIS3L1 a | XRN1 a | DIS3L2 b |
| SMG5 | ↑ | ↓ | ↔ | ↔ | ↑ | ↔ |
| SMG1 | ↑ | ↔ | ↔ | ↔ | ↔ | ↑ |
| SLC7A11 | ↑ | ↑ | ↑ | ↑ | ↔ | ↑ |
| GADD45A | ↑ | ↔ | ↔ | ↔ | ↔ | ↑ |
| GADD45B | ↑ | ↔ | ↔ | ↔ | ↔ | ↑ |
| GABARAPL1 | ↑ | ↓ | ↑ | ↔ | ↑ | ↔ |
| ANTXR1 | ↑ | ↔ | ↔ | ↔ | ↔ | ↔ |
| SLC1A3 | ↑ | ↑ | ↑ | ↑ | ↔ | ↑ |
| PLXNA1 | ↔ | ↔ | ↔ | ↔ | ↑ | ↑ |
| ATF3 | ↔ | ↔ | ↔ | ↔ | ↔ | ↑ |
| ARFRP1 | ↑ | ↔ | ↔ | ↔ | ↔ | ↔ |
| BAG1 | ↑ | ↔ | ↔ | ↔ | ↔ | ↔ |
| Targets | SMG6 | PM/Scl100 | DIS3 | DIS3L1 | XRN1 |
|---|---|---|---|---|---|
| βWT | ↑ | ↑ | ↑ | ↑ | ↑ |
| β15 | ↑ | ↑ | ↑ | ↑ | ↑ |
| β26 | ↑ | ↔ | ↑ | ↔ | ↔ |
| β39 | ↑ | ↔ | ↑ | ↔ | ↔ |
| βNS | ↔ | ↑ | ↑ | ↔ | ↔ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa, P.J.; Menezes, J.; Guedes, R.; Reis, F.P.; Teixeira, A.; Saramago, M.; Viegas, S.C.; Arraiano, C.M.; Romão, L. A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay. Genes 2024, 15, 1308. https://doi.org/10.3390/genes15101308
da Costa PJ, Menezes J, Guedes R, Reis FP, Teixeira A, Saramago M, Viegas SC, Arraiano CM, Romão L. A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay. Genes. 2024; 15(10):1308. https://doi.org/10.3390/genes15101308
Chicago/Turabian Styleda Costa, Paulo J., Juliane Menezes, Raquel Guedes, Filipa P. Reis, Alexandre Teixeira, Margarida Saramago, Sandra C. Viegas, Cecília M. Arraiano, and Luísa Romão. 2024. "A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay" Genes 15, no. 10: 1308. https://doi.org/10.3390/genes15101308
APA Styleda Costa, P. J., Menezes, J., Guedes, R., Reis, F. P., Teixeira, A., Saramago, M., Viegas, S. C., Arraiano, C. M., & Romão, L. (2024). A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay. Genes, 15(10), 1308. https://doi.org/10.3390/genes15101308





