Identification of Putative Quantitative Trait Loci for Improved Seed Oil Quality in Peanuts
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Conditions
2.2. Sample Preparation and Fatty Acid Analysis
2.3. Statistical Analysis
2.4. QTL Mapping Analysis
3. Results
3.1. Detection and Variation of Major Fatty Acids in the Seeds of the Two Parental Genotypes
3.2. Phenotypic Variations among Recombinant Inbred Lines
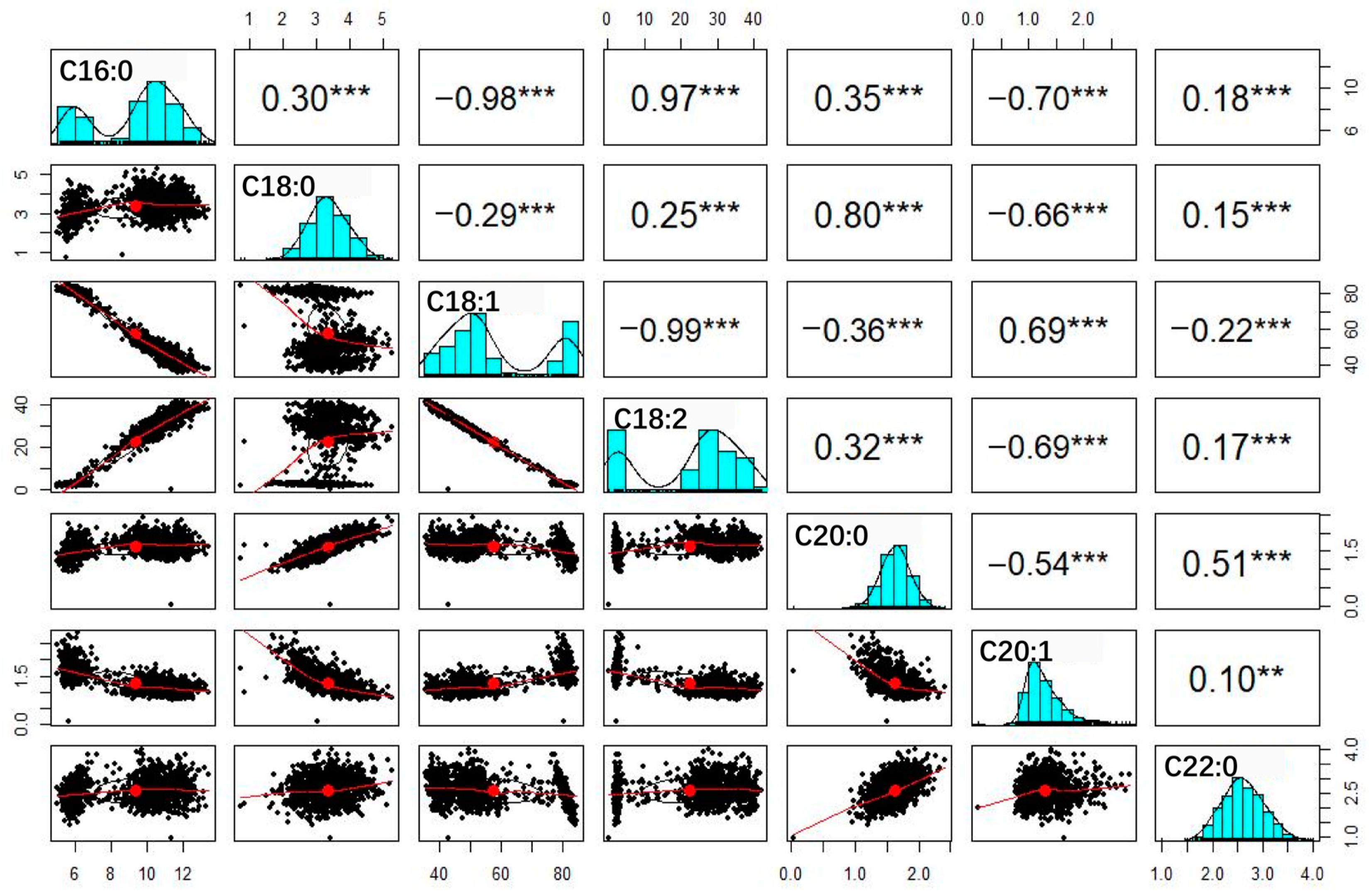
3.3. Correlation Analysis of Fatty Acid Traits
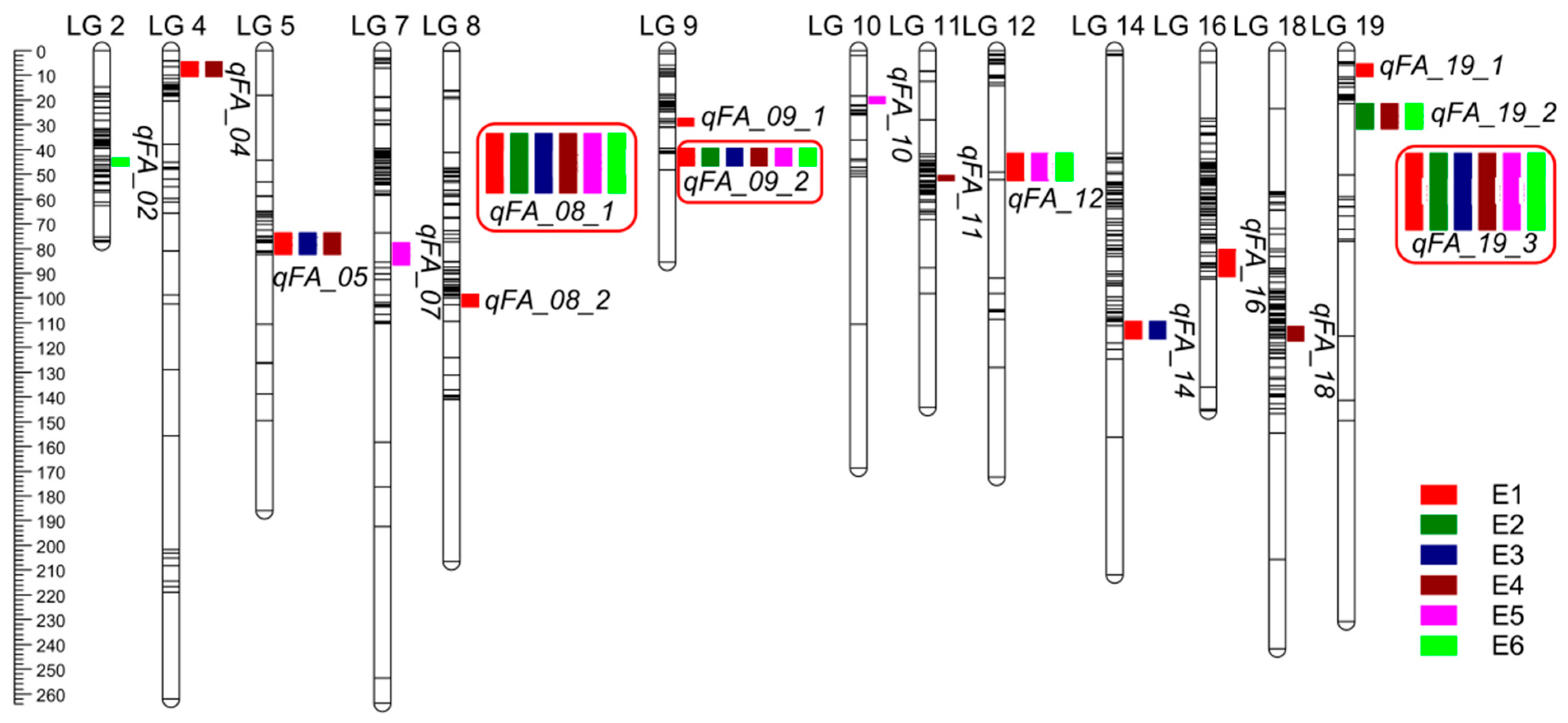
3.4. QTL Analysis of Fatty Acids in Individual Environments
3.5. Joint Analysis of Fatty Acid Traits in Multiple Environments
3.6. Detection of Stable Epistatic QTL in Individual-Environment Trials
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kavera, B. Genetic improvement for oil quality through induced mutagenesis in groundnut (Arachis hypogaea L.). Ph.D. Thesis, University of Agricultural Sciences, Dharwad, India, 2008. [Google Scholar]
- Mahboubifar, M.; Yousefinejad, S.; Alizadeh, M.; Hemmateenejad, B. Prediction of the acid value, peroxide value and the percentage of some fatty acids in edible oils during long heating time by chemometrics analysis of FTIR-ATR spectra. J. Iran. Chem. Soc. 2016, 13, 2291–2299. [Google Scholar] [CrossRef]
- Lawrence, G.D. Perspective: The saturated fat–unsaturated oil dilemma: Relations of dietary fatty acids and serum cholesterol, atherosclerosis, inflammation, cancer, and all-cause mortality. Adv. Nutr. 2021, 12, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Su, C. Fatty Acid Composition of Oils, Their Oxidative, Flavor and Heat Stabilities and the Resultant Quality in Foods. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2003. [Google Scholar]
- Vassiliou, E.; Gonzalez, A.; Garcia, C.; Tadros, J.; Toney, J. Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-α both in vitro and in vivo systems. Lipids Health Dis. 2009, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Qiu, X.; Hu, C.; Li, J.; Wu, L.; Wang, W. Breeding of a new variety of peanut with high-oleic-acid content and high-yield by marker-assisted back-crossing. Mol. Breed. 2022, 42, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yoo, E.; Lee, S.; Sung, J.; Noh, H.J.; Wang, S.J.; Desta, K.T. Seed weight and genotype influence the total oil content and fatty acid composition of peanut seeds. Foods 2022, 11, 3463. [Google Scholar] [CrossRef] [PubMed]
- Singkham, N.; Jogloy, S.; Kesmala, T.; Swatsitang, P.; Jaisil, P.; Puppala, N. Genotypic variability and genotype by environment interactions in oil and fatty acids in high, intermediate, and low oleic acid peanut genotypes. J. Agric. Food Chem. 2010, 58, 6257–6263. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.L.; Nigam, S.C.N.; Rao, R.C.; Singh, U.; Rao, K.V.S. Effect of drought on oil, fatty acids and protein contents of groundnut (Arachis hypogaea L.) seeds. Field Crops Res. 1996, 48, 125–133. [Google Scholar] [CrossRef]
- Golombek, S.D.; Sridhar, R.; Singh, U. Effect of Soil Temperature on the seed composition of three spanish cultivars of groundnut (Arachis hypogaea L.). J. Agric. Food Chem. 1995, 43, 2067–2070. [Google Scholar] [CrossRef]
- Pandey, M.K.; Guo, B.; Holbrook, C.C.; Janila, P.; Zhang, X.; Bertioli, D.J.; Isobe, S.; Liang, S.; Varshney, R.K. Molecular markers, genetic maps and QTL for molecular breeding in peanut. (Arachis hypogaea L.). BMC Genet. 2014, 61–113. [Google Scholar]
- Bilgrami, S.; Liu, L.; Farokhzadeh, S.; Najafabadi, A.S.; Ramandi, H.D.; Nasiri, N.; Darwish, I. Meta-analysis of QTL controlling seed quality traits based on QTL alignment in brassica napus. Ind. Crops Prod. 2022, 176, 114307. [Google Scholar] [CrossRef]
- Pandey, M.K.; Wang, M.L.; Qiao, L.; Feng, S.; Khera, P.; Wang, H.; Tonnis, B.; Barkley, N.A.; Wang, J.P.; Holbrook, C.C. Identification of QTL associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.). BMC Genet. 2014, 15, 133. [Google Scholar] [CrossRef] [PubMed]
- Addisu, G.H.; Yang, X.L.; He, M.J. Peanuts, genetic map construction and QTL mapping research progress. J. Peanuts 2017, 46, 10. (In English) [Google Scholar]
- Cheng, L.Q.; Wang, J.; Huang, L. Molecular markers in genetic mapping and QTL mapping of peanut. J. Peanuts 2016, 45, 5. [Google Scholar]
- Zhang, X.Y. Genetic Analysis and QTL Mapping of Yield, Quality and Disease Resistance of Cultivated Peanut. Ph.D. Thesis, Zhejiang University, Hangzhou, China, 2011. [Google Scholar]
- Sun, Z.; Qi, F.; Liu, H.; Qin, L.; Xu, J.; Shi, L.; Zhang, Z.; Miao, L.; Huang, B.; Dong, W. QTL mapping of quality traits in peanut using whole-genome resequencing. Crop J. 2022, 10, 177–184. [Google Scholar] [CrossRef]
- Wilson, J.N.; Baring, M.R.; Burow, M.D.; Rooney, W.L.; Chagoya, J.C. Generation means analysis of fatty acid composition in peanut. J. Crop Improv. 2013, 27, 430–443. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Li, Y.R.; Cheng, Z.S.; Su, Q.; Jin, X.X.; Song, Y.H.; Wang, J. Genetic analysis and exploration of major effect QTL underlying oil content in peanut. Theor. Appl. Genet. 2023, 136, 97. [Google Scholar] [CrossRef]
- Wang, M.L.; Khera, P.; Pandey, M.K.; Hui, L. Genetic mapping of QTL controlling fatty acids provided insights into the genetic control of fatty acid synthesis pathway in peanut (Arachis hypogaea L.). PLoS ONE 2015, 10, e0119454. [Google Scholar] [CrossRef]
- George, D.; Mallery, P. IBM SPSS Statistics 19 Step by Step a Simple Guide and Reference; Pearson: London, UK, 2013. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTL. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Bera, S.K.; Kamdar, J.; Thankappan, R. High-oleic peanut in India: Shift in paradigm of health oil. In Proceedings of the XIV Agricultural Science Congress, New Delhi, India, 20–23 February 2019. [Google Scholar]
- Pattee, H.E.; Isleib, T.G.; Moore, K.M.; Moore, K.M.; Gorbet, D.W.; Giesbrecht, F.G. Effect of high-oleic trait and paste storage variables on sensory attribute stability of roasted peanuts. J. Agric. Food Chem. 2002, 50, 7366–7370. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, M.L.; Barkley, N.A.; Pittman, R.N. A simple allele-specific PCR assay for detecting FAD2 alleles in both A and B genomes of the cultivated peanut for high-oleate trait selection. Plant Mol. Biol. Rep. 2010, 28, 542–548. [Google Scholar] [CrossRef]
- Yu, H.T.; Yang, W.Q.; Tang, Y.Y.; Wang, X.Z.; Wu, Q.; Hu, D.Q.; Wang, C.T.; Yu, S.L. An AS-PCR assay for accurate genotyping of FAD2A/FAD2B genes in peanuts (Arachis hypogaea L.). Grasas Aceites 2013, 64, 395–399. [Google Scholar]
- Nawade, B.; Bosamia, T.C.; Thankappan, R.; Arulthambi, R.L.; Abhay, K.; Jentilal, D.R.; Rahul, K.; Gyan, M.P. Insights into the Indian peanut genotypes for ahFAD2 gene polymorphism regulating its oleic and linoleic acid fluxes. Front. Plant Sci. 2016, 7, 1271. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Sun, J.; Sun, J.; Zhang, X.; Zhao, C.; Pan, J.; Hou, L.; Tian, R.; Wang, X. Insights into the novel FAD2 gene regulating oleic acid accumulation in peanut seeds with different maturity. Genes 2022, 13, 2076. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.R.; Cheng, Z.S.; Chen, S.L.; Song, Y.H.; Liu, Y.J.; Zhang, P.J. Identification of ahFAD gene variation and genetic diversity analysis of peanut varieties (lines) with high oleic acid. Journal of Plant Genetic Resources. 2020, 21, 208–214. [Google Scholar]
- Kochert, G.; Stalker, H.T.; Gimenes, M.A.; Galgaro, M.L.; Lopes, C.R.; Moore, K. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae). Am. J. Bot. 1996, 83, 1282–1291. [Google Scholar] [CrossRef]
- Seijo, J.G.; Lavia, G.I.; Fernández, A.; Krapovickas, A.; Ducasse, D.; Moscone, E.A. Physical mapping of the 5s and 18s–25s rRNA genes by FISH as evidence that Arachis duranensis and A. ipaensis are the wild diploid progenitors of A. hypogaea (Leguminosae). Am. J. Bot. 2004, 91, 1294–1303. [Google Scholar] [CrossRef]


| Traits | Parents | RILs | H2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| JH5 | KX01-6 | Mean | SD | CV% | Max | Min | Kurt | Skew | ||
| C16:0 | 11.61 | 5.72 | 9.32 | 2.27 | 24.32 | 13.40 | 4.94 | −1.09 | −0.55 | 0.986 |
| C18:0 | 4.24 | 2.07 | 3.36 | 0.60 | 17.82 | 5.29 | 0.75 | 0.35 | 0.01 | 0.932 |
| C18:1 | 42.02 | 80.87 | 57.71 | 15.36 | 26.62 | 84.78 | 35.51 | −1.13 | 0.61 | 0.987 |
| C18:2 | 35.72 | 3.33 | 22.57 | 13.14 | 58.21 | 41.97 | 0.41 | −1.16 | −0.58 | 0.987 |
| C20:0 | 1.87 | 1.16 | 1.63 | 0.23 | 14.26 | 2.42 | 0.04 | 1.75 | −0.26 | 0.921 |
| C20:1 | 0.91 | 2.28 | 1.29 | 0.33 | 25.69 | 2.82 | 0.08 | 2.06 | 1.25 | 0.966 |
| C22:0 | 2.42 | 2.66 | 2.61 | 0.43 | 16.34 | 4.01 | 0.99 | 0.00 | 0.17 | 0.876 |
| Fatty Acid | Group | C18:0 | C20:0 | C20:1 | C22:0 |
|---|---|---|---|---|---|
| C16:0 | Peak1 | 0.34 *** | 0.31 *** | −0.16 ** | 0.28 *** |
| Peak2 | −0.07 * | −0.05 ns | −0.24 *** | 0.00 ns | |
| C18:1 | Peak1 | −0.31 *** | 0.47 *** | −0.08 ns | −0.58 *** |
| Peak2 | 0.12 *** | 0.03 ns | 0.17 *** | −0.09 ** | |
| C18:2 | Peak1 | −0.01 ns | 0.01 ns | 0.03 ns | 0.07 ns |
| Peak2 | −0.21 *** | −0.10 ** | −0.18 *** | 0.01 ns |
| QTL | Fatty Acid | LG | Environment | Position | Left Marker | Right Marker | LOD | PVE% | Add | Threshold |
|---|---|---|---|---|---|---|---|---|---|---|
| qFA_02 | C20:0 | 2 | E6 | 44 | Chr02.92004332 | Chr02.92097568 | 4.63 | 4.28 | 0.05 | 4.20 |
| qFA_04 | C18:0, C20:0 | 4 | E1, E4 | 7–8 | Chr04.7737038 | Chr04.10252892 | 4.48–5.90 | 5.60–6.90 | 0.05–0.15 | 4.20–4.40 |
| qFA_05 | C20:0 | 5 | E1, E3, E4 | 75–82 | Chr05.37478237 | Chr05.88563212 | 6.13–31.03 | 11.63–12.40 | 0.07–0.33 | 4.50–8.00 |
| qFA_07 | C22:0 | 7 | E5 | 83 | Chr07.4150878 | Chr07.3251821 | 4.71 | 8.50 | 0.10 | 4.30 |
| qFA_08_1 | C18:0, C20:0, C20:1 | 8 | All | 39–55 | Chr08.42575311 | Chr08.46776535 | 6.33–15.58 | 9.08–22.43 | 0.10–0.29 | 4.20–6.60 |
| qFA_08_2 | C22:0 | 8 | E1 | 100 | Chr08.15237857 | Chr08.14658317 | 5.48 | 6.14 | 0.08 | 4.80 |
| qFA_09_1 | C18:0 | 9 | E1 | 28 | Chr09.111222696 | Chr09.111444555 | 9.35 | 11.57 | 0.17 | 4.40 |
| qFA_09_2 | C16:0, C18:0, C18:1, C18:2, C20:0, C20:1 | 9 | All | 40–41 | Chr09.113696781 | Chr09.114431570 | 5.12–32.93 | 6.37–28.79 | 0.05–9.65 | 4.00–6.60 |
| qFA_10 | C18:2 | 10 | E5 | 19 | Chr10.5820536 | Chr10.6895328 | 4.23 | 3.21 | 2.72 | 4.20 |
| qFA_11 | C22:0 | 11 | E4 | 51 | Chr11.135590338 | Chr11.134662327 | 5.35 | 9.89 | 0.09 | 4.50 |
| qFA_12 | C20:0, C22:0 | 12 | E1, E5, E6 | 49–50 | Chr12.17574379 | Chr12.3375707 | 5.59–9.80 | 6.11–17.13 | 0.04–0.11 | 4.30–4.70 |
| qFA_14 | C20:0 | 14 | E1, E3 | 110 | Chr14.139524574 | Chr14.137319755 | 5.14–18.38 | 5.52–11.80 | 0.04–0.16 | 4.30–13.30 |
| qFA_16 | C20:0 | 16 | E1 | 89 | Chr16.147123562 | Chr16.146960299 | 6.18 | 6.69 | 0.05 | 4.30 |
| qFA_18 | C18:0 | 18 | E4 | 112 | Chr18.114667904 | Chr18.115066398 | 5.58 | 6.97 | 0.16 | 4.20 |
| qFA_19_1 | C18:2 | 19 | E1 | 7 | Chr19.8567191 | Chr19.11838675 | 4.12 | 2.57 | 2.51 | 4.10 |
| qFA_19_2 | C16:0, C18:1, C18:2 | 19 | E2, E4, E6 | 11–20 | Chr19.12239093 | Chr19.12852116 | 4.02–10.14 | 2.64–7.50 | 0.47–5.27 | 4.00–4.10 |
| qFA_19_3 | C16:0, C18:1, C18:2, C20:0, C20:1 | 19 | All | 39–46 | Chr19.138748208 | Chr19.156112831 | 5.99–37.56 | 9.14–38.72 | 0.07–11.35 | 4.00–6.60 |
| QTL | LG | Trait | Position | Marker Interval | LOD | PVE% | Add | Interaction Effect between Additive QTL and Environment | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A% | AE% | AE1 | AE2 | AE3 | AE4 | AE5 | AE6 | |||||||
| qFA_02 | 2 | C20:0 | 44 | Chr02.92004332_Chr02.92097568 | 6.40 | 1.04 | 1.07 | 1.50 | −0.06 | −0.06 | −1.74 | −1.41 | 0.30 | 2.96 |
| qFAm_04 | 4 | C20:1 | 17 | Chr04.87381676_Chr04.80904224 | 38.42 | 4.14 | 4.67 | −7.16 | 7.26 | −15.79 | 3.91 | −1.36 | 5.20 | 0.79 |
| qFA_05 | 5 | C22:0 | 75 | Chr05.90854275_Chr05.88563212 | 42.81 | 10.99 | 20.52 | 8.53 | 2.66 | −4.53 | 24.71 | −6.35 | −8.44 | −8.05 |
| 5 | C20:0 | 78 | Chr05.41888224_Chr05.37841097 | 11.34 | 2.16 | 2.76 | 2.16 | −2.07 | 0.62 | −2.23 | 4.16 | −2.32 | 1.83 | |
| qFA_08_1 | 8 | C18:0 | 42 | Chr08.46776535_Chr08.44595184 | 16.76 | 3.94 | 4.57 | 7.75 | −5.56 | −2.39 | −5.57 | 18.37 | −2.14 | −2.71 |
| qFA_09_1 | 9 | C18:0 | 28 | Chr09.111222696_Chr09.111444555 | 11.26 | 1.86 | 2.11 | 5.32 | 12.09 | −5.57 | −1.90 | −3.29 | −0.69 | −0.63 |
| qFA_12 | 12 | C22:0 | 49 | Chr12.17574379_Chr12.3375707 | 19.29 | 2.43 | 4.86 | −4.01 | −6.93 | 3.96 | 4.06 | 3.99 | −9.02 | 3.94 |
| qFAm_16_3 | 16 | C22:0 | 87 | Chr16.147687093_Chr16.147523996 | 6.70 | 1.08 | 1.25 | −2.94 | −5.20 | −3.32 | 3.82 | 1.66 | 1.83 | 1.20 |
| qFA_16 | 16 | C20:0 | 89 | Chr16.147123562_Chr16.146960299 | 8.93 | 1.07 | 1.59 | −1.62 | −2.84 | −1.48 | 3.50 | 0.01 | −0.09 | 0.90 |
| Epistatic QTL | Environment | Trait | Loc.1 | Pos.1 | Marker interval | Loc.2 | Pos.2 | Marker interval | LOD | PVE% | Add1 | Add2 | Add by Add |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qFA_E8 | E1, E3 | C16:0, C18:1 | 8 | 170 | Chr08.2179124_Chr08.43274117 | 9 | 40 | Chr09.113696781_ Chr09.115494682 | 5.00–6.66 | 4.51–7.32 | 0.67–5.02 | 1.22–8.84 | 0.67–3.96 |
| qFA_E9_1 | E1, E3 | C16:0, C18:1, C18:2 | 9 | 40 | Chr09.113696781_ Chr09.115494682 | 9 | 70 | Chr09.115494715_Chr09.112209867 | 5.24–9.30 | 4.65–13.79 | 1.20–8.93 | 0.51–3.74 | 0.84–6.13 |
| qFA_E9_2 | E1, E3 | C16:0, C18:1, C18:2 | 9 | 40 | Chr09.113696781_ Chr09.115494682 | 10 | 80 | Chr10.20573531_Chr10.2981198 | 5.01–8.87 | 4.55–13.99 | 1.21–8.82 | 0.54–3.93 | 0.82–5.37 |
| qFA_E9_3 | E1, E2, E3, E4, E5 | C16:0, C18:1, C18:2, C20:1 | 9 | 40 | Chr09.113696781_ Chr09.115494682 | 19 | 35 | Chr19.138748208_Chr19.156112831 | 5.92–28.01 | 8.02–34.97 | 0.00–9.57 | 0.13–10.95 | 0.09–4.30 |
| qFA_E9_4 | E2, E4, E5, E6 | C16:0, C18:1, C18:2 | 9 | 45 | Chr09.113696781_Chr09.115494682 | 9 | 70 | Chr09.115494715_Chr09.112209867 | 5.10–8.43 | 5.21–9.02 | 0.10–0.83 | 0.50–4.17 | 0.87–5.85 |
| qFA_E9_5 | E4, E6 | C16:0, C18:1, C18:2 | 9 | 45 | Chr09.113696781_Chr09.115494682 | 10 | 80 | Chr10.20573531_Chr10.2981198 | 5.29–6.41 | 5.94–7.64 | 0.00–0.14 | 0.70–4.84 | 0.76–5.63 |
| qFA_E9_6 | E5, E6 | C16:0, C18:1, C18:2 | 9 | 45 | Chr09.113696781_Chr09.115494682 | 12 | 145 | Chr12.3561394_Chr12.116523714 | 5.05–5.74 | 4.54–6.82 | 0.22–0.87 | 0.62–4.04 | 0.75–5.08 |
| qFA_E9_7 | E4, E5, E6 | C16:0, C18:1, C18:2 | 9 | 45 | Chr09.113696781_Chr09.115494682 | 13 | 155 | Chr13.143629587_Chr13.138922790 | 5.15–5.89 | 4.91–6.64 | 0.04–0.33 | 0.71–5.05 | 0.73–4.61 |
| qFA_E10 | E1, E3 | C16:0, C18:1, C18:2 | 10 | 80 | Chr10.20573531_Chr10.2981198 | 19 | 145 | Chr19.155910347_Chr19.153836481 | 5.29–7.25 | 1.40–4.10 | 0.56–4.04 | 0.12–0.89 | 0.78–5.21 |
| qFA_E14 | E1, E4 | C16:0, C18:1, C20:1 | 14 | 190 | Chr14.13558383_Chr14.5680674 | 19 | 145 | Chr19.155910347_Chr19.153836481 | 5.05–6.64 | 1.30–9.28 | 0.06–4.23 | 0.04–0.36 | 0.13–4.36 |
| qFA_E19 | E1, E3, E4, E5, E6 | C16:0, C18:1, C18:2, C20:1 | 19 | 40 | Chr19.138748208_Chr19.156112831 | 19 | 145 | Chr19.155910347_Chr19.153836481 | 6.54–29.13 | 4.98–25.01 | 1.49–11.07 | 0.00–0.83 | 0.09–4.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, P.; Zhang, J.; Song, Y.; Zhao, X.; Jin, X.; Su, Q.; Yang, Y.; Wang, J. Identification of Putative Quantitative Trait Loci for Improved Seed Oil Quality in Peanuts. Genes 2024, 15, 75. https://doi.org/10.3390/genes15010075
Hu P, Zhang J, Song Y, Zhao X, Jin X, Su Q, Yang Y, Wang J. Identification of Putative Quantitative Trait Loci for Improved Seed Oil Quality in Peanuts. Genes. 2024; 15(1):75. https://doi.org/10.3390/genes15010075
Chicago/Turabian StyleHu, Pengju, Jianan Zhang, Yahui Song, Xing Zhao, Xinxin Jin, Qiao Su, Yongqing Yang, and Jin Wang. 2024. "Identification of Putative Quantitative Trait Loci for Improved Seed Oil Quality in Peanuts" Genes 15, no. 1: 75. https://doi.org/10.3390/genes15010075
APA StyleHu, P., Zhang, J., Song, Y., Zhao, X., Jin, X., Su, Q., Yang, Y., & Wang, J. (2024). Identification of Putative Quantitative Trait Loci for Improved Seed Oil Quality in Peanuts. Genes, 15(1), 75. https://doi.org/10.3390/genes15010075






