Genome-Wide Association Analysis of Reproductive Traits in Chinese Holstein Cattle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Genotype and Quality Control
2.2. Statistical Analysis
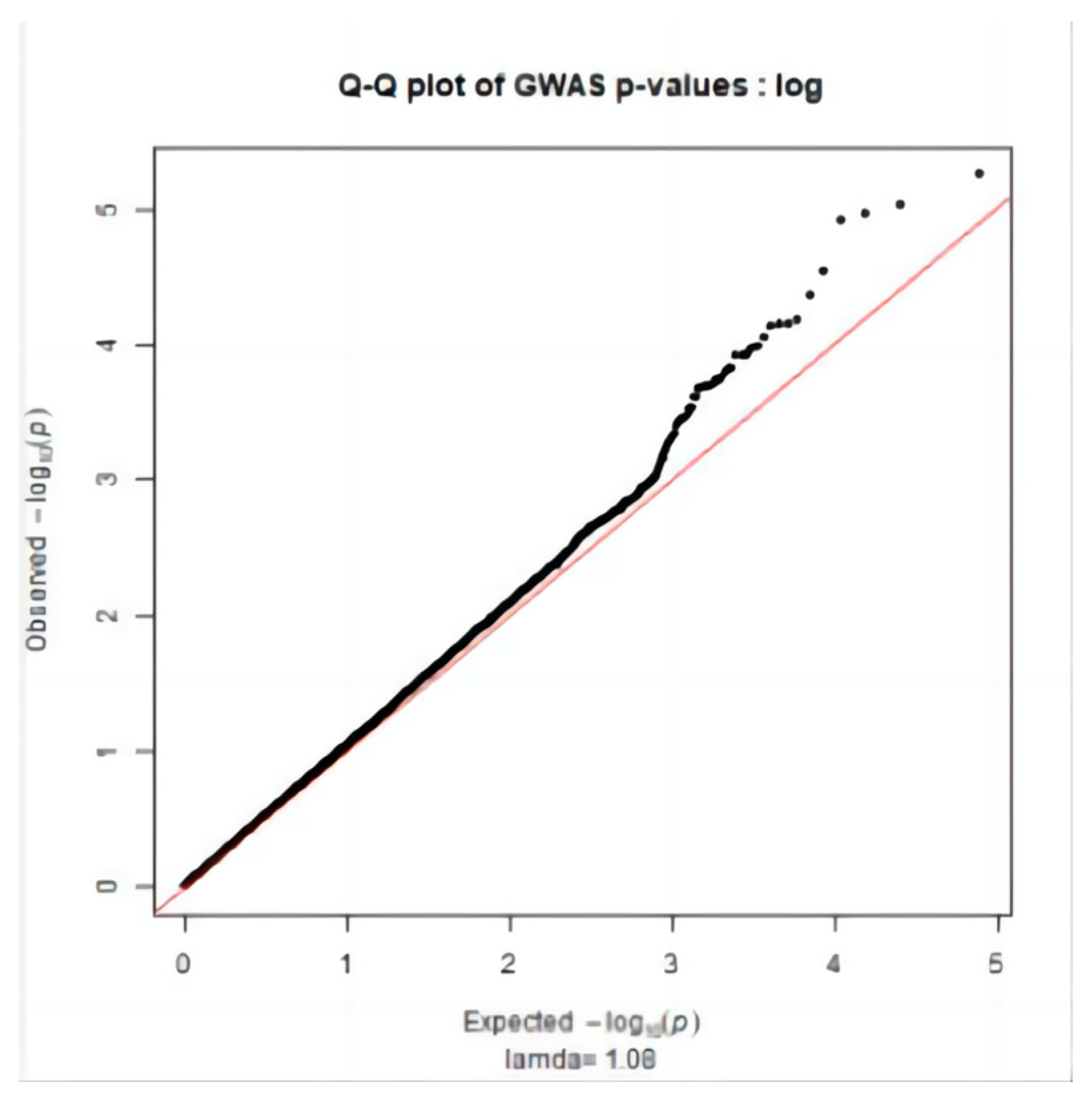
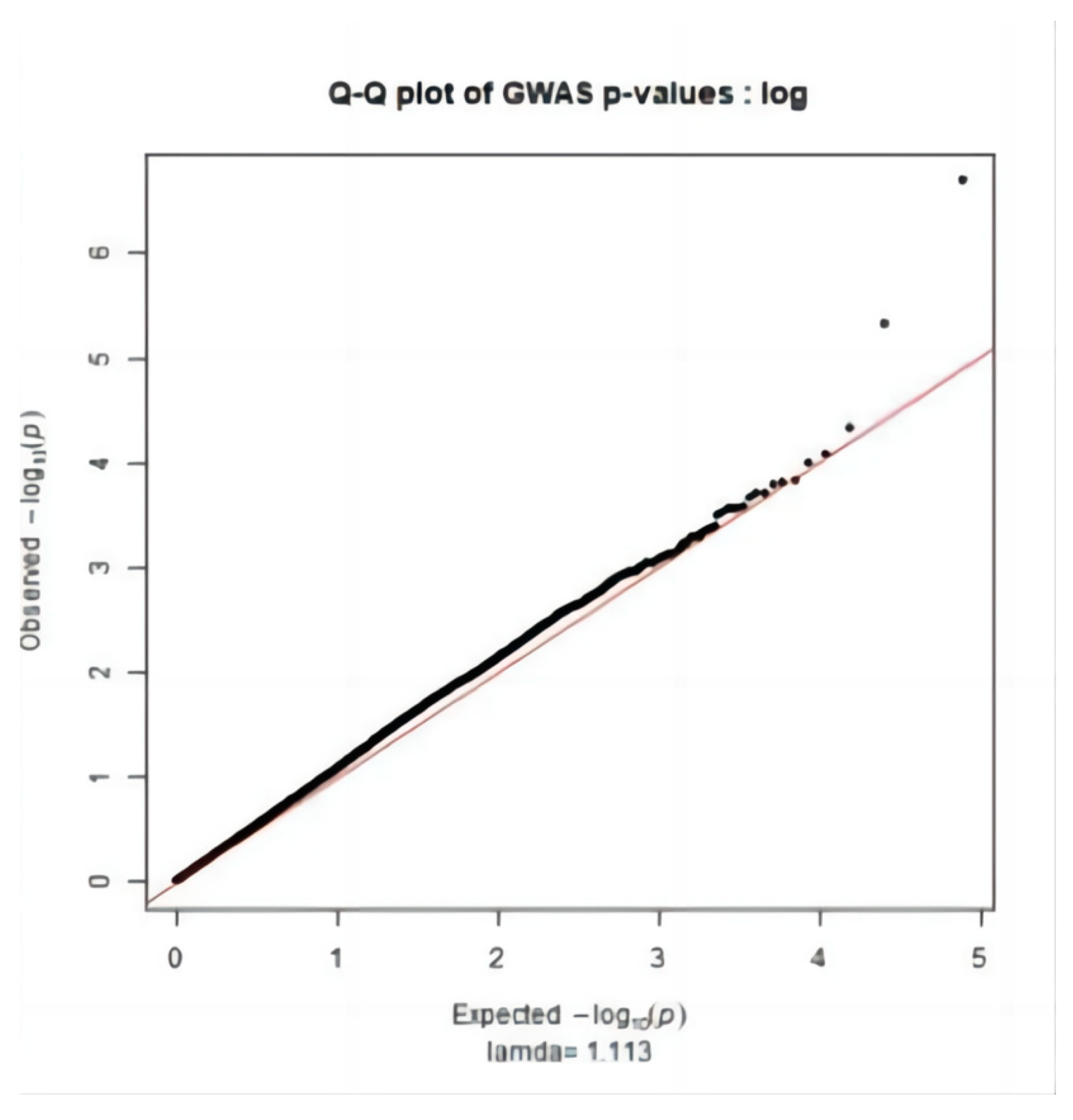
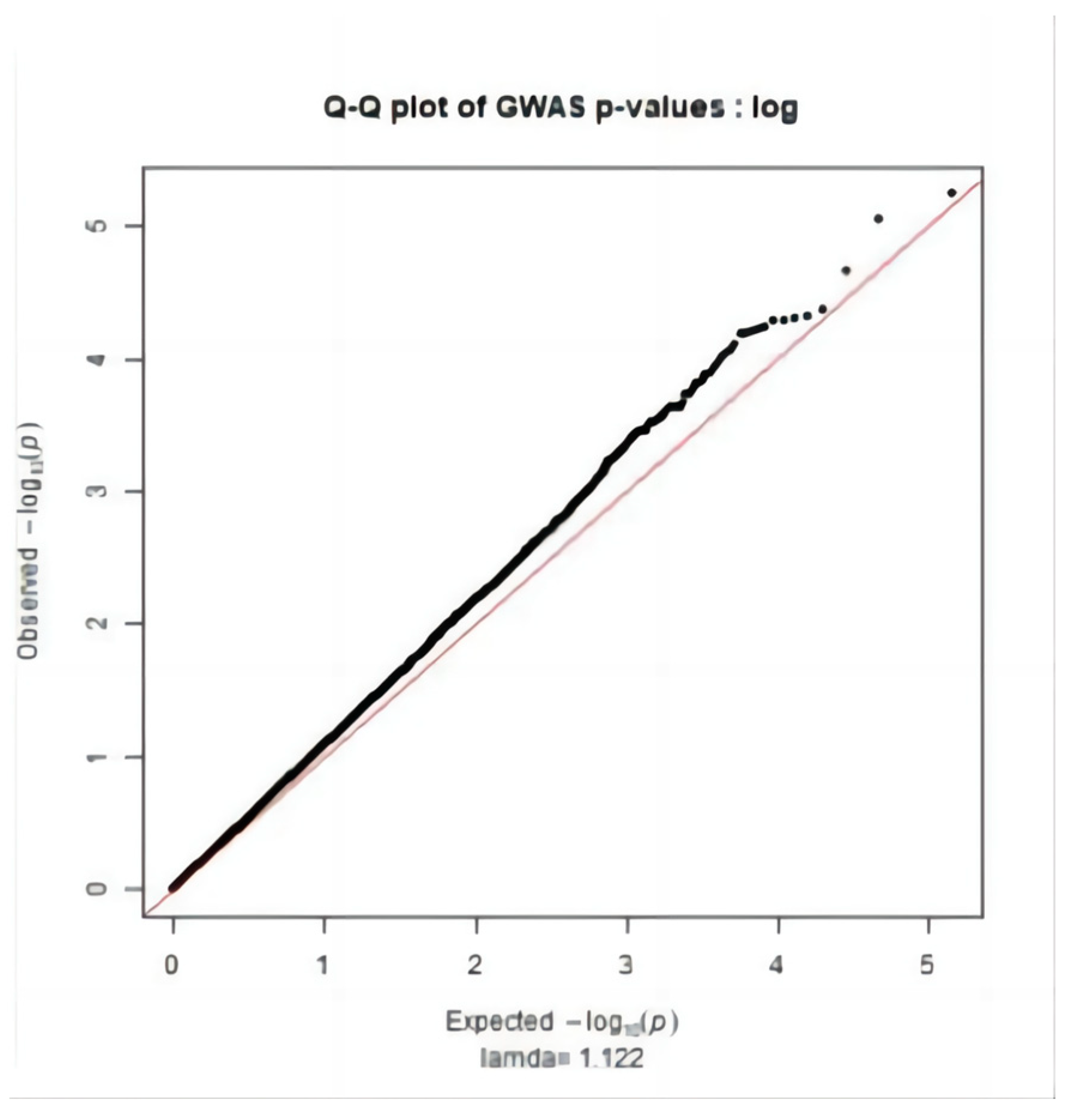
2.3. Significance Test
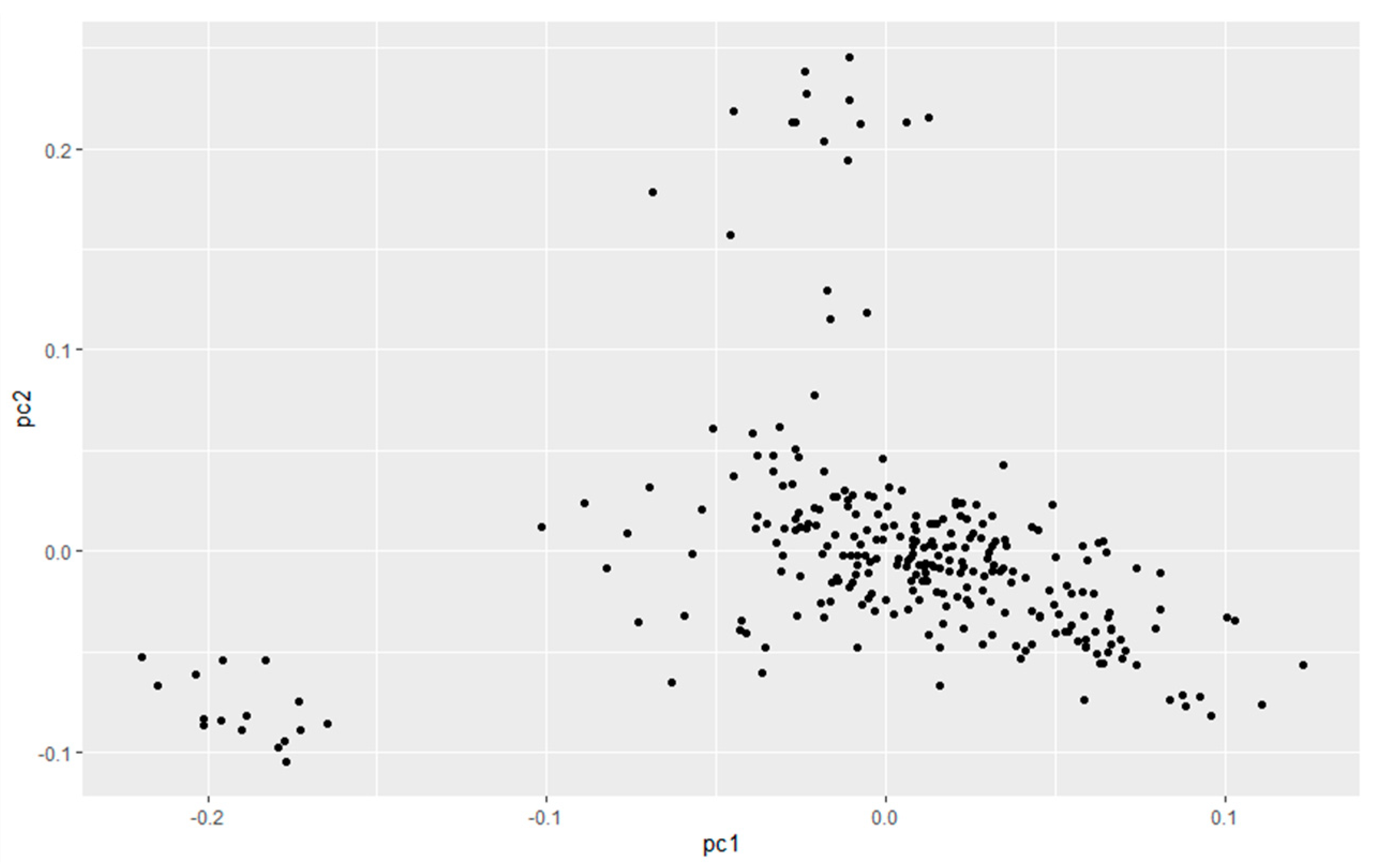
2.4. Population Stratification Analysis
2.5. Screening and Annotation of the Candidate Genes
2.6. Enrichment Analysis of the Candidate Genes
2.7. Software and the Database Website Used in This Study
3. Results
3.1. Population Stratification
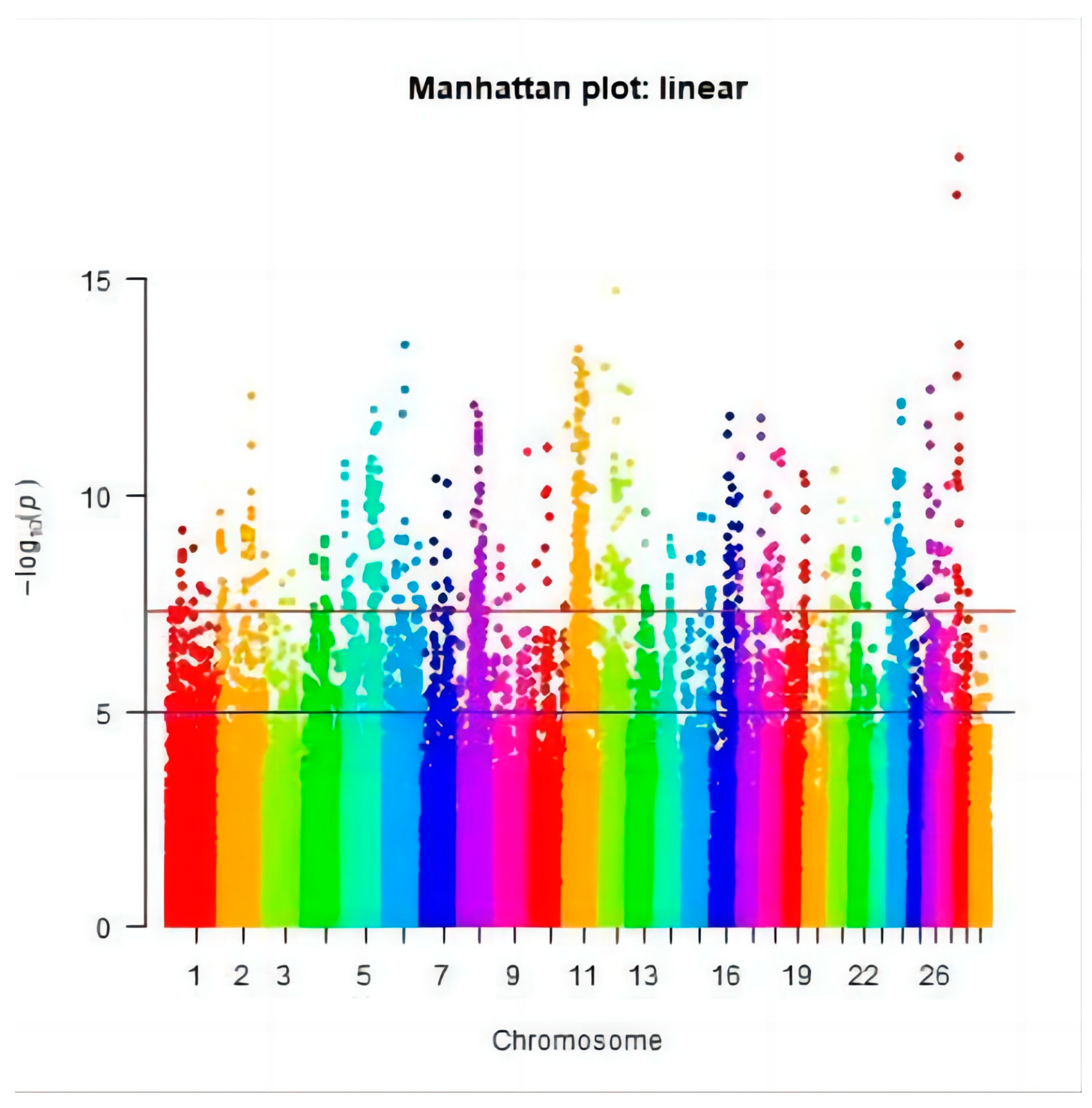
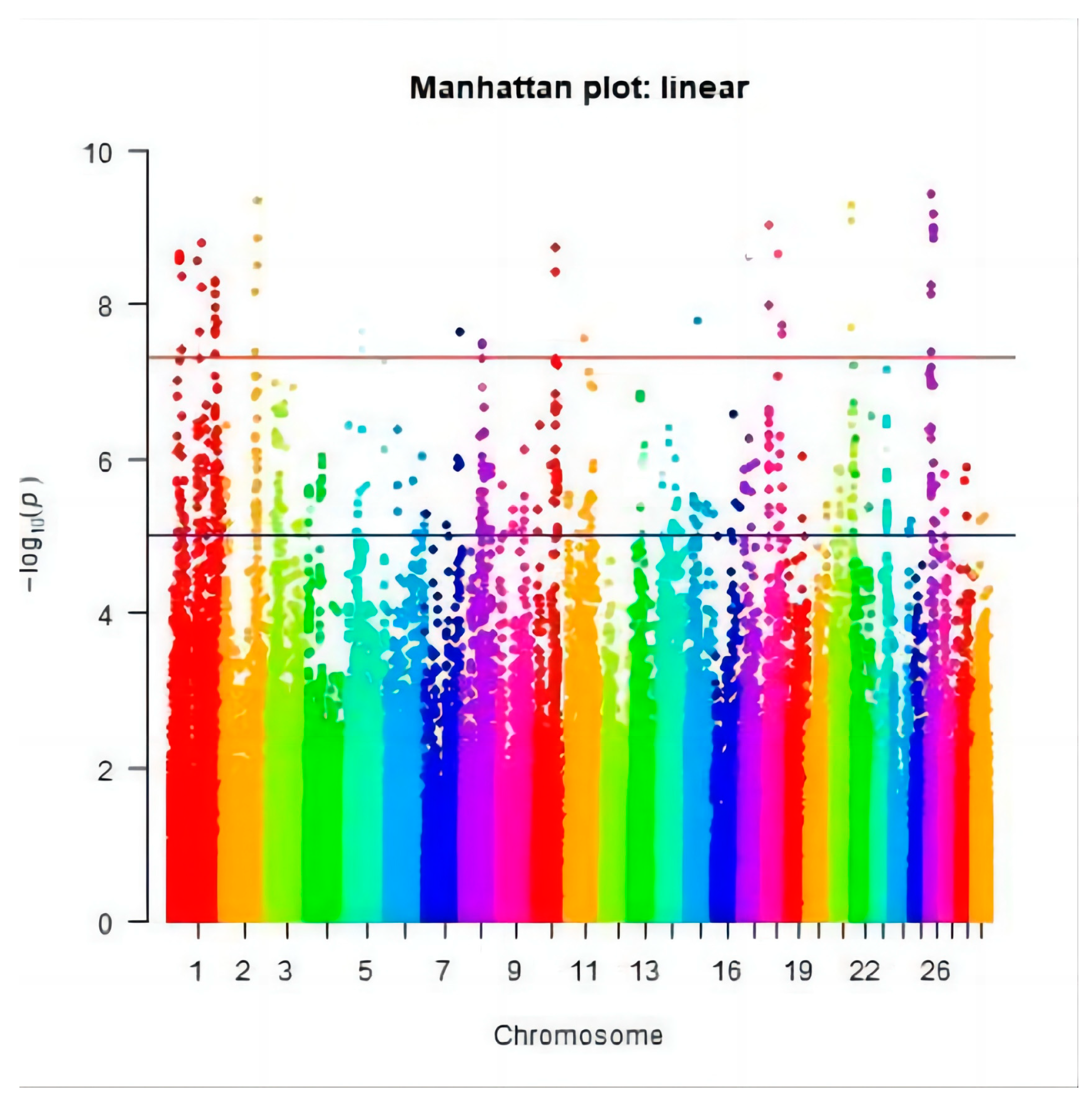
3.2. Genome-Wide Association Study
3.3. Significant SNPs and the Candidate Genes
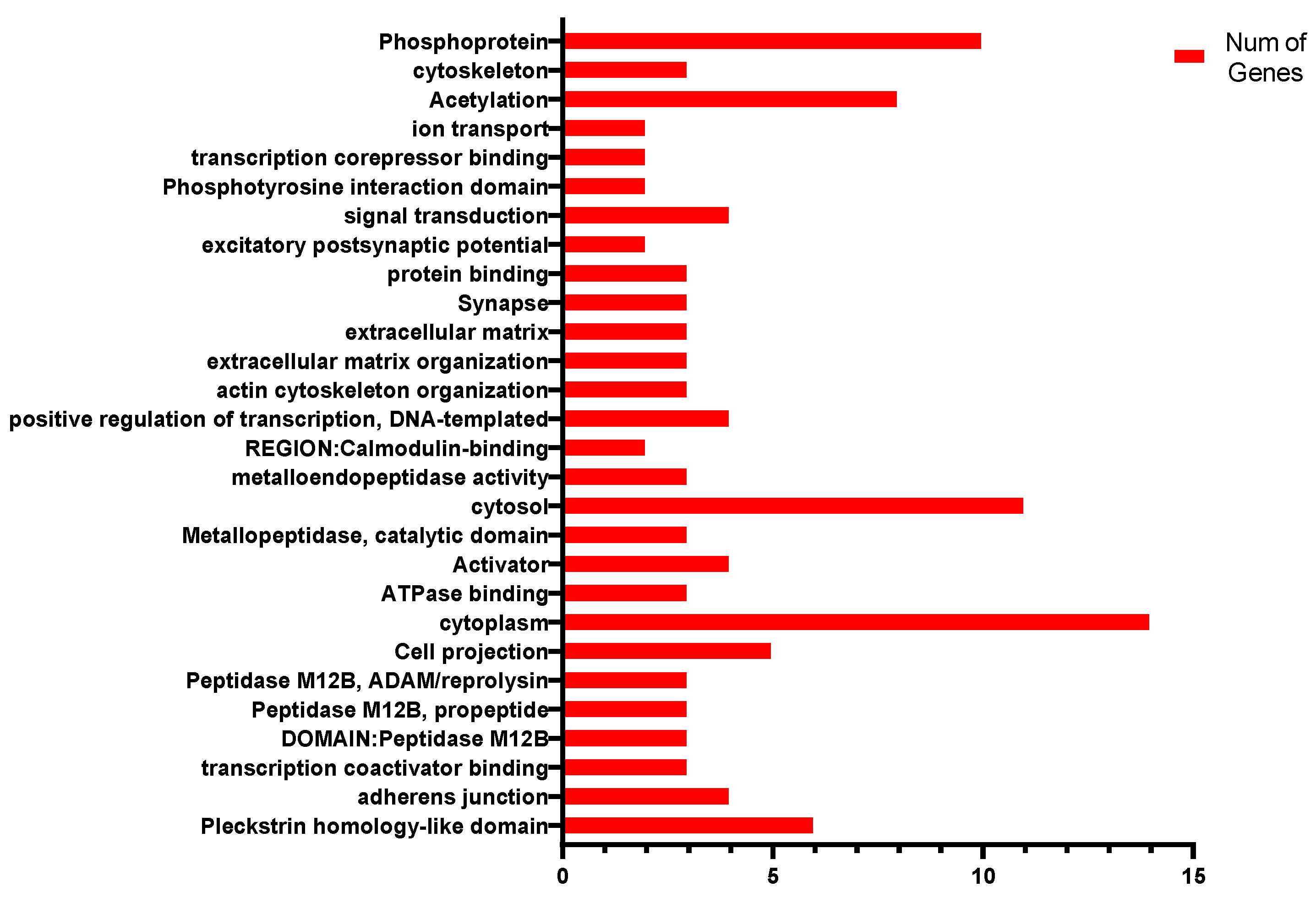
3.4. Enrichment Analysis of the Candidate Genes
4. Discussion
4.1. Genome-Wide Association Study
4.1.1. Candidate Genes Associated with the HCR
4.1.2. Candidate Genes Associated with the CCR
4.1.3. Candidate Genes Associated with the DPR
4.2. Candidate-Gene Enrichment Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Risch, N.; Merikangas, K. The Future of Genetic Studies of Complex Human Diseases. Science 1996, 273, 1516–1517. [Google Scholar] [CrossRef] [PubMed]
- Iniguez_Luy, H.A.G.B. Association mapping of seed quality traits inBrassica napusl using GWAS and candidate QTL approaches. Mol. Breed. 2015, 35, 143. [Google Scholar]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Hermas SA Young, C.W.; Rust, J.W. Genetic Relationships and Additive Genetic Variation of Productive and Reproductive Traits in Guernsey Dairy Cattle. J. Dairy Sci. 1987, 70, 1252–1257. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Muhammad Suhail, S.; Shafiq, M. Heritability estimates and genetic correlations of various production and reproductive traits of different grades of dairy cattle realed under subtropical condition. Reprod. Domest. Anim. 2019, 54, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Olsen, H.G.; Hayes, B.J.; Kent, M.P.; Nome, T.; Svendsen, M.; Lien, S. A genome wide association study for QTL affecting direct and maternal effects of stillbirth and dystocia in cattle. Anim. Genet. 2010, 41, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Sahana, G.; Guldbrandtsen, B.; Thomsen, B.; Lund, M.S. Confirmation and fine-mapping of clinical mastitis and somatic cell score QTL in Nordic Holstein cattle. Anim. Genet. 2013, 44, 620–626. [Google Scholar] [CrossRef]
- Hering, D.; Olenski, K.; Kaminski, S. Genome-wide association study for poor sperm motility in Holstein-Friesian bulls. Anim. Reprod. Sci. 2014, 146, 89–97. [Google Scholar] [CrossRef]
- Huang, W.; Kirkpatrick, B.W.; Rosa, G.J.M.; Khatib, H. A genome-wide association study using selective DNA pooling identifies candidate markers for fertility in Holstein cattle. Anim. Genet. 2010, 41, 570–578. [Google Scholar] [CrossRef]
- Liu, A.; Guo, G.; Wang, Y. Genetic assessment and genome-wide association analysis of Holstein cattle in China. J. Anim. Husb. Vet. Med. 2015, 46, 373–381. [Google Scholar]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Weedon, M.N.; Purcell, S.; Lettre, G.; Estrada, K.; Willer, C.J.; Visscher, P.M. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 2011, 19, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, J.; Sun, D.; Ma, P.; Ding, X.; Yu, Y.; Zhang, Q. Genome Wide Association Studies for Milk Production Traits in Chinese Holstein Population. PLoS ONE 2010, 5, e13661. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gao, X.; Song, W.; Yao, D.; Chen, L.; Chen, C.; Ma, Y. Genome-wide association analysis of limb-hoof structure and breast morphology in Chinese Holstein cattle. J. Livest. Ecol. 2021, 42, 13–18. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association Mapping in Structured Populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.S.; Barratt, B.J.; Clayton, D.G.; Todd, J.A. Genome-wide association studies: Theoretical and practical concerns. Nat. Rev. Genet. 2005, 6, 109–118. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A. Why Most Published Research Findings Are False. PLoS Med. 2005, 2, e124. [Google Scholar] [CrossRef]
- Li, Y. Genome-Wide Association Analysis of Biochemical Components of Blood in Chinese Holstein Dairy Cows. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2020. [Google Scholar]
- Vilhjálmsson, B.J.; Nordborg, M. The nature of confounding in genome-wide association studies. Nat. Rev. Genet. 2013, 14, 1–2. [Google Scholar] [CrossRef]
- Kim, S.; Xing, E.P. Statistical estimation of correlated genome associations to a quantitative traits network. PLoS Genet. 2009, 5, e1000587. [Google Scholar] [CrossRef]
- O’Reilly, P.F.; Hoggart, C.J.; Pomyen, Y.; Calboli, F.C.; Elliott, P.; Jarvelin, M.R.; Coin, L.J. Multi-Phen: Joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE 2012, 7, e34861. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, Y.; Katafuchi, M.; Almehmadi, A.; Venkitapathi, S.; Jaha, H.; Ehrenman, J.; Morcos, J.; Aljamaan, R.; Mochida, Y. Modulation of matrix mineralization by Vwc2-like protein and its novel splicing isoforms. Biochem. Biophys. Res. Commun. 2012, 418, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, D.; Hernandez-Sanchez, J.; Chen, J.; Liu, C.; Wu, Z.; Fang, M.; Li, N. Genome Wide Association Analysis Reveals New Production Trait Genes in a Male Duroc Population. PLoS ONE 2015, 10, e0139207. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.V.; Eozenou, C.; Healey, G.D.; Forde, N.; Reinaud, P.; Chebrout, M.; Gall, L.; Rodde, N.; Padilla, A.L.; Delville, C.G.; et al. Analysis of STAT1 expression and biological activity reveals interferon-tau-dependent STAT1-regulated SOCS genes in the bovine endometrium. Reprod. Fertil. Dev. 2016, 28, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Li, J.; Zhu, H.; Liu, J.; Dong, S.; Li, L.; Qu, L.; Chen, H.; Song, X.; Lan, X. Deletion mutation within the goat PPP3CA gene identified by GWAS significantly affects litter size. Reprod. Fertil. Dev. 2021, 33, 476–483. [Google Scholar] [CrossRef]
- Pathak, P.; Blech-Hermoni, Y.; Subedi, K.; Mpamugo, J.; Obeng-Nyarko, C.; Ohman, R.; Molloy, I.; Kates, M.; Hale, J.; Stauffer, S.; et al. Myopathy associated LDB3 mutation causes Z-disc disassembly and protein aggregation through PKCα and TSC2-mTOR downregulation. Commun. Biol. 2021, 4, 355. [Google Scholar] [CrossRef]
- van Dijk, M.; Mulders, J.; Könst, A.; Janssens, B.; van Roy, F.; Blankenstein, M.; Oudejans, C. Differential downregulation of αT-catenin expression in placenta: Trophoblast cell type-dependent imprinting of the CTNNA3 gene. Gene Expr. Patterns 2004, 5, 61–65. [Google Scholar] [CrossRef]
- Tai, C.S.; Chen, Y.Y.; Chen, W.L. β-Lactoglobulin Influences Human Immunity and Promotes Cell Proliferation. BioMed Res. Int. 2016, 2016, 7123587. [Google Scholar] [CrossRef]
- Kämäräinen, M.; Riittinen, L.; Seppälä, M.; Palotie, A.; Andersson, L.C. Progesterone-associated endometrial protein--a constitutive marker of human erythroid precursors. Blood 1994, 84, 467–473. [Google Scholar] [CrossRef]
- Sbardella, A.P.; Watanabe, R.N.; da Costa, R.M.; Bernardes, P.A.; Braga, L.G.; Baldi Rey, F.S.; Lôbo, R.B.; Munari, D.P. Genome-Wide Association Study Provides Insights into Important Genes for Reproductive Traits in Nelore Cattle. Animals 2021, 11, 1386. [Google Scholar] [CrossRef]
- Zeng, F.; Tian, Y.; Shi, S.; Wu, Q.; Liu, S.; Zheng, H.; Yue, L.; Li, Y. Identification of mouse MARVELD1 as a microtubule associated protein that inhibits cell cycle progression and migration. Mol. Cells 2011, 31, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Shimba, S.; Wada, T.; Hara, S.; Tezuka, M. EPAS1 Promotes Adipose Differentiation in 3T3-L1 Cells. J. Biol. Chem. 2004, 279, 40946–40953. [Google Scholar] [CrossRef] [PubMed]
- Baguma-Nibasheka, M.; Fracassi, A.; Costain, W.J.; Moreno, S.; Kablar, B. Role of skeletal muscle in motor neuron development. Histol. Histopathol. 2016, 31, 699–719. [Google Scholar] [PubMed]
- Zhou, D.; Wang, Y.; Yang, R.; Wang, F.; Zhao, Z.; Wang, X.; Xie, L.; Tian, X.; Wang, G.; Li, B.; et al. The MyoD1 Promoted Muscle Differentiation and Generation by Activating CCND2 in Guanling Cattle. Animals 2022, 12, 2571. [Google Scholar] [CrossRef] [PubMed]
- Geyer, J.; Döring, B.; Failing, K.; Petzinger, E. Molecular cloning and functional characterization of the bovine (Bos taurus) organic anion transporting polypeptide Oatp1a2 (Slco1a2). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 137, 317–329. [Google Scholar] [CrossRef]
- Wild-Bode, C.; Fellerer, K.; Kugler, J.; Haass, C.; Capell, A. A Basolateral Sorting Signal Directs ADAM10 to Adherens Junctions and Is Required for Its Function in Cell Migration. J. Biol. Chem. 2006, 281, 23824–23829. [Google Scholar] [CrossRef]
- Yang, Z.; Cool, B.H.; Martin, G.M.; Hu, Q. A Dominant Role for FE65 (APBB1) in Nuclear Signaling. J. Biol. Chem. 2006, 281, 4207–4214. [Google Scholar] [CrossRef]
- Ortega, M.S.; Wohlgemuth, S.; Tribulo, P.; Siqueira, L.G.B.; Null, D.J.; Cole, J.B.; Da Silva, M.V.; Hansen, P.J. A single nucleotide polymorphism in COQ9 affects mitochondrial and ovarian function and fertility in Holstein cows. Biol. Reprod. 2017, 96, 652–663. [Google Scholar] [CrossRef]

| Trait | Number | Average | Maximum | Minimum | Variance | Standard Deviation |
|---|---|---|---|---|---|---|
| HCR | 637 | 0.81 | 4.1 | −2.4 | 1.30 | 1.14 |
| CCR | 637 | 0.62 | 4.9 | −3.1 | 1.69 | 1.30 |
| DPR | 637 | 0.00031 | 3.4 | −3.2 | 1.27 | 1.13 |
| Before Quality Control | After Quality Control | ||||
|---|---|---|---|---|---|
| Chromosome | Length (Mb) | SNPs Count | Density (kb/snp) | SNPs Count | Density (kb/snp) |
| 1 | 158.3 | 5556 | 35.1 | 4693 | 29.6 |
| 2 | 137.1 | 4688 | 34.2 | 3917 | 28.6 |
| 3 | 121.4 | 4508 | 37.1 | 3749 | 30.9 |
| 4 | 120.8 | 4049 | 33.5 | 3390 | 28.1 |
| 5 | 121.2 | 4523 | 37.3 | 3734 | 30.8 |
| 6 | 119.5 | 4364 | 36.5 | 3617 | 30.3 |
| 7 | 112.6 | 3903 | 34.7 | 3232 | 28.7 |
| 8 | 113.4 | 3805 | 33.6 | 3289 | 29.0 |
| 9 | 105.7 | 3695 | 35.0 | 3181 | 30.1 |
| 10 | 104.3 | 3626 | 34.8 | 3116 | 29.9 |
| 11 | 107.3 | 3801 | 35.4 | 3216 | 30.0 |
| 12 | 91.2 | 3044 | 33.4 | 2563 | 28.1 |
| 13 | 84.2 | 3064 | 36.4 | 2617 | 31.1 |
| 14 | 84.6 | 3045 | 36.0 | 2561 | 30.3 |
| 15 | 85.3 | 3119 | 36.6 | 2674 | 31.3 |
| 16 | 81.7 | 2826 | 34.6 | 2338 | 28.6 |
| 17 | 75.2 | 2668 | 35.5 | 2286 | 30.4 |
| 18 | 66.0 | 2605 | 39.5 | 2197 | 33.3 |
| 19 | 64.1 | 2726 | 42.5 | 2226 | 34.7 |
| 20 | 72.0 | 2737 | 38.0 | 2253 | 31.3 |
| 21 | 71.6 | 2573 | 35.9 | 2167 | 30.3 |
| 22 | 61.4 | 2201 | 35.8 | 1878 | 30.6 |
| 23 | 52.5 | 2110 | 40.2 | 1774 | 33.4 |
| 24 | 62.7 | 2259 | 36.0 | 1934 | 30.8 |
| 25 | 42.9 | 1726 | 40.2 | 1443 | 33.6 |
| 26 | 51.7 | 1823 | 35.3 | 1551 | 30.0 |
| 27 | 45.4 | 1699 | 37.4 | 1488 | 32.8 |
| 28 | 46.3 | 1735 | 37.5 | 1506 | 32.5 |
| 29 | 51.5 | 1871 | 36.3 | 1570 | 30.5 |
| Character | SNP | Chromosome | Site (bp) | p Value | The Gene Region | Distance (bp) |
|---|---|---|---|---|---|---|
| HCR | BovineHD0200029628 | 2 | 102,699,078 | 9.18 × 10−3 | VWC2L | 81,620 |
| BovineHD0200022967 | 2 | 79,537,846 | 3.11 × 10−5 | STAT1 | 21,534 | |
| BTA-37416-no-rs | 6 | 23,807,252 | 8.69 × 10−4 | PPP3CA | 36,965 | |
| BovineHD28000743 | 28 | 41,387,894 | 7.36 × 10−5 | LDB3 | 2984 | |
| BovineHD2800005891 | 28 | 22,263,728 | 6.23 × 10−5 | CTNNA3 | 10,962 | |
| CCR | LGB_X14710_5174 | 11 | 103,259,143 | 7.68 × 10−3 | PAEP | 1719 |
| BovineHD1100000394 | 11 | 1,230,012 | 3.72 × 10−4 | ACOXL | 6654 | |
| BovineHD4100008487 | 11 | 2,827,428 | 2.86 × 10−4 | EPAS1 | 73,158 | |
| BovineHD1700011952 | 17 | 42,103,930 | 4.12 × 10−4 | GLRB | 6465 | |
| BovineHD2600004843 | 26 | 18,941,126 | 4.61 × 10−5 | MARVELD1 | 68,791 | |
| DPR | ARS-USMARC-666 | 5 | 25,527,614 | 1.51 × 10−6 | PDE1B | 11,893 |
| BovineHD0500025349 | 5 | 88,911,724 | 6.17 × 10−4 | SLCO1A2 | 22,340 | |
| BovineHD0700016212 | 7 | 54,281,313 | 6.66 × 10−4 | ARHGAP26 | 3543 | |
| Hapmap58695-rs29019899 | 10 | 51,673,875 | 1.95 × 10−5 | ADAM10 | 5385 | |
| BovineHD1500013531 | 15 | 46,637,606 | 1.29 × 10−5 | APBB1 | 11,182 | |
| UFL-rs41859871 | 18 | 4,419,224 | 1.94 × 10−5 | MON1B | 3866 | |
| COQ9_rs109301586 | 18 | 25,446,323 | 4.27 × 10−4 | COQ9 | 2139 | |
| ARS-BFGL-NGS-115062 | 21 | 67,739,884 | 8.39 × 10−3 | CDC42BPB | 18,754 | |
| ARS-BFGL-NGS-115980 | 26 | 20,196,747 | 1.74 × 10−5 | HPSE2 | 61,344 | |
| BovineHD2600004843 | 26 | 18,941,126 | 4.61 × 10−5 | MARVELD1 | 68,791 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Xu, L.; Ding, X.; Ma, Y. Genome-Wide Association Analysis of Reproductive Traits in Chinese Holstein Cattle. Genes 2024, 15, 12. https://doi.org/10.3390/genes15010012
Liu J, Xu L, Ding X, Ma Y. Genome-Wide Association Analysis of Reproductive Traits in Chinese Holstein Cattle. Genes. 2024; 15(1):12. https://doi.org/10.3390/genes15010012
Chicago/Turabian StyleLiu, Jiashuang, Lingyang Xu, Xiangbin Ding, and Yi Ma. 2024. "Genome-Wide Association Analysis of Reproductive Traits in Chinese Holstein Cattle" Genes 15, no. 1: 12. https://doi.org/10.3390/genes15010012
APA StyleLiu, J., Xu, L., Ding, X., & Ma, Y. (2024). Genome-Wide Association Analysis of Reproductive Traits in Chinese Holstein Cattle. Genes, 15(1), 12. https://doi.org/10.3390/genes15010012




