Genetic Characterization of Blood Group Antigens for Polynesian Heritage Norfolk Island Residents
Abstract
1. Introduction
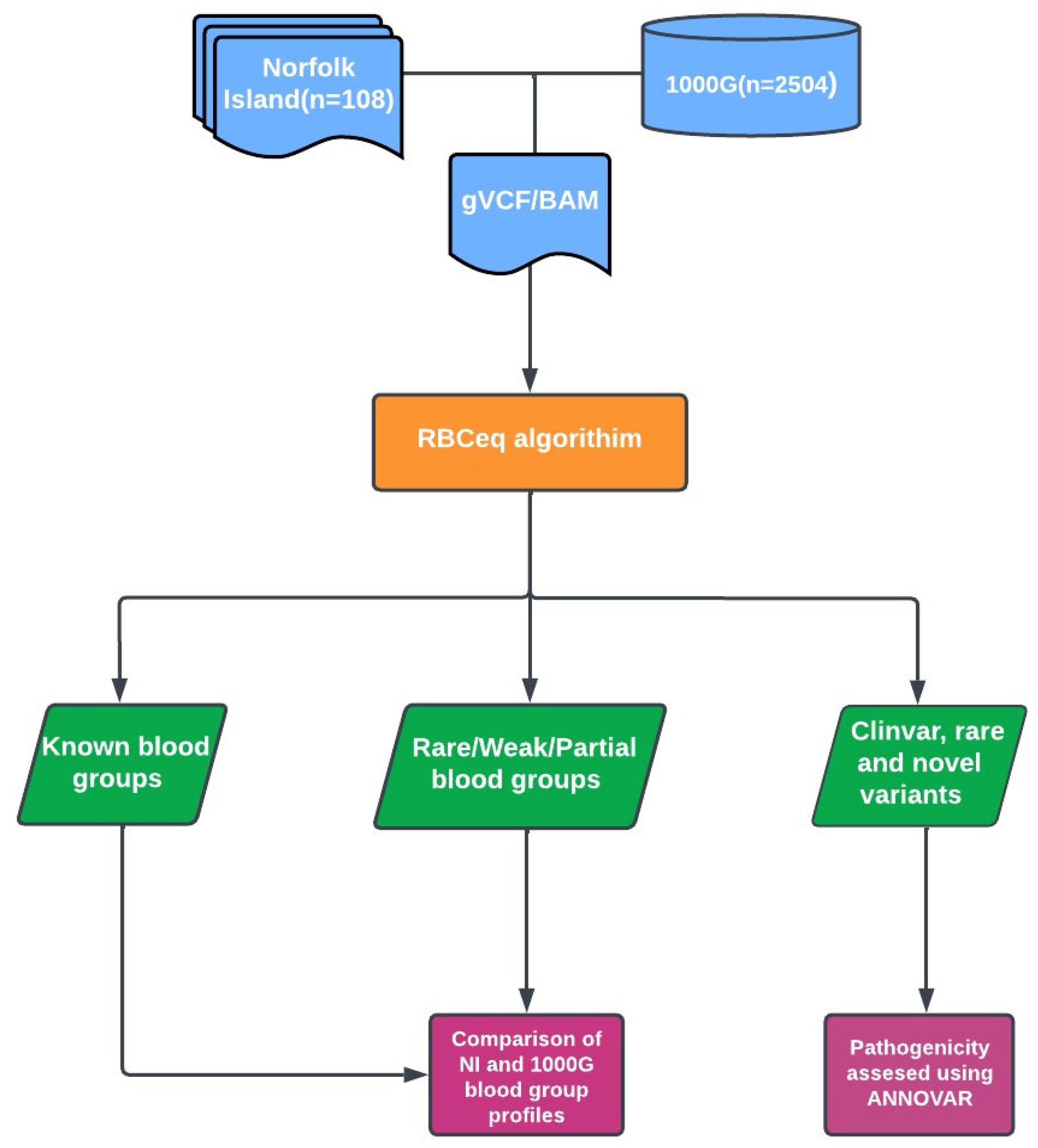
2. Methods
2.1. Norfolk Island Population Sample Selection and Whole Genome Sequencing Data
2.2. Variant Calling and Annotation
2.3. Prediction of Red Cell Genotypes and Phenotypes Using RBCeq
2.4. Copy Number Variation Analysis
2.5. Comparison with 1000 Genomes (1000G) Dataset
2.6. Statistical Analysis
3. Results
3.1. Prevalence of Predicted ABO Phenotype
3.2. RHD Characterisation and Its Prevalence
3.3. Comprehensive Analysis of the Kidd Blood Group among Norfolk Islanders
3.4. Prediction and Distribution of High Frequency Antigens
3.5. Analysis of Copy Number Variations among Norfolk Island Residents
3.6. Prevalence of Predicted Weak Phenotypes and Low Frequency Antigens among Norfolk Island Residents
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ajmani, P.S. Blood Group and Immunology. In Immunohematology and Blood Banking: Principles and Practice; Springer: Singapore, 2020; pp. 7–23. [Google Scholar]
- Harmening, D.M. Modern Blood Banking & Transfusion Practices; FA Davis: Philadelphia, PA, USA, 2018. [Google Scholar]
- Connie, M.; Westhoff, J.R.S.; Beth, H.S. Chapter 110—Human Blood Group Antigens and Antibodies. In Human Blood Group Antigens and Antibodies, 7th ed.; Hoffman, E.J.B.R., Silberstein, L.E., Heslop, H.E., Weitz, J.I., Anastasi, J., Salama, M.E., Abutalib, S.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 1687–1701. [Google Scholar]
- Karpinski, M.; Pochinco, D.; Dembinski, I.; Laidlaw, W.; Zacharias, J.; Nickerson, P. Leukocyte reduction of red blood cell transfusions does not decrease allosensitization rates in potential kidney transplant candidates. Am. Soc. Nephrol. 2004, 15, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Gill, K.S.; Muntner, P.; Lafayette, R.A.; Petersen, J.; Fink, J.C.; Gilbertson, D.T.; Bradbury, B.D. Red blood cell transfusion use in patients with chronic kidney disease. Nephrol. Dial. Transplant. 2013, 28, 1504–1515. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caamaño, J.; Musante, E.; Contreras, M.; Ulloa, H.; Reyes, C.; Inaipil, V.; Saavedra, N.; Guzmán, N. Frequency and specificity of red blood cell alloimmunization in chilean transfused patients. Transfus. Med. Hemotherapy 2015, 42, 4–7. [Google Scholar] [CrossRef] [PubMed]
- ISBT. Table of Blood Group Systems v10.0. ISBT: Red Cell Immunogenetics and Blood Group Terminology. 2021. Available online: https://www.isbtweb.org/fileadmin/user_upload/Table_of_blood_group_systems_v10.0_30-JUN-2021_with_LRG_and_revised_antigens.pdf (accessed on 11 May 2023).
- Keller, M. The role of red cell genotyping in transfusion medicine. Immunohematology 2015, 31, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Jadhao, S.; Hoy, W.; Lee, S.; Patel, H.R.; McMorran, B.J.; Flower, R.L.; Nagaraj, S.H. The genomic landscape of blood groups in Indigenous Australians in remote communities. Transfusion 2022, 62, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Hatzikotoulas, K.; Gilly, A.; Zeggini, E. Using population isolates in genetic association studies. Brief. Funct. Genom. 2014, 13, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Australian Bureau of Statistics. Norfolk Island. Norfolk Island 2016 Census All Persons QuickStats. 2016. Available online: https://www.abs.gov.au/census/find-census-data/quickstats/2016/SSC90004 (accessed on 22 June 2022).
- Macgregor, S.; Bellis, C.; Lea, R.A.; Cox, H.; Dyer, T.; Blangero, J.; Visscher, P.M.; Griffiths, L.R. Legacy of mutiny on the Bounty: Founder effect and admixture on Norfolk Island. Eur. J. Hum. Genet. 2010, 18, 67–72. [Google Scholar] [CrossRef]
- Lawicki, S.; Covin, R.B.; Powers, A.A. The Kidd (JK) blood group system. Transfus. Med. Rev. 2017, 31, 165–172. [Google Scholar] [CrossRef]
- KPMG. Norfolk Island Health Service Plan. 2020. Available online: https://www.regional.gov.au/territories/norfolk_island/norfolk_island_health_services/files/2019-kpmg-ni-health-service-plan.pdf (accessed on 22 June 2022).
- Australian Bureau of Statistics. 3301.0-Births, Australia, 2016. Available online: https://www.abs.gov.au/AUSSTATS/abs@.nsf/Lookup/3301.0Explanatory%20Notes12016?OpenDocument (accessed on 22 June 2022).
- Benton, M.C. Identification of Susceptibility Genes for Metabolic Syndrome in the Isolated Population of Norfolk Island. Ph.D. Thesis, Griffith University, Queensland, Australia, 2013. [Google Scholar]
- Bellis, C.; Hughes, R.M.; Begley, K.N.; Quinlan, S.; Lea, R.A.; Heath, S.C.; Blangero, J.; Griffiths, L.R. Phenotypical characterisation of the isolated Norfolk Island population focusing on epidemiological indicators of cardiovascular disease. Hum. Hered. 2005, 60, 211–219. [Google Scholar] [CrossRef]
- Tervonen, H.; Foliaki, S.; Bray, F.; Roder, D. Cancer epidemiology in the small nations of Pacific Islands. Cancer Epidemiol. 2017, 50, 184–192. [Google Scholar] [CrossRef]
- Park, H.; Liu, X.; Henry, L.; Harman, J.; Ross, E.A. Trends in anemia care in non-dialysis-dependent chronic kidney disease (CKD) patients in the United States (2006–2015). BMC Nephrol. 2018, 19, 318. [Google Scholar] [CrossRef] [PubMed]
- Docherty, A.B.; Walsh, T.S. Anemia and blood transfusion in the critically ill patient with cardiovascular disease. Crit. Care 2017, 21, 61. [Google Scholar] [CrossRef] [PubMed]
- National Blood Authority Australia. National Blood Authority Annual Report 2019–2020. 2020. Available online: www.blood.gov.au/about-nba (accessed on 9 May 2022).
- Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; del Angel, G.; Rivas, M.A.; Hanna, M.; McKenna, A.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Jadhao, S.; Davison, C.L.; Roulis, E.V.; Schoeman, E.M.; Divate, M.; Haring, M.; Williams, C.; Shankar, A.J.; Lee, S.; Pecheniuk, N.M.; et al. RBCeq: A robust and scalable algorithm for accurate genetic blood typing. EBioMedicine 2022, 76, 103759. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Geoffroy, V.; Herenger, Y.; Kress, A.; Stoetzel, C.; Piton, A.; Dollfus, H.; Muller, J. AnnotSV: An integrated tool for structural variations annotation. Bioinformatics 2018, 34, 3572–3574. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Muñoz, E.A.; Milko, L.V.; Harrison, S.M.; Azzariti, D.R.; Kurtz, C.L.; Lee, K.; Mester, J.L.; Weaver, M.A.; Currey, E.; Craigen, W.; et al. ClinGen variant curation expert panel experiences and standardized processes for disease and gene-level specification of the ACMG/AMP guidelines for sequence variant interpretation. Hum. Mutat. 2018, 39, 1614–1622. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef]
- NHS. Blood Groups. 2020. Available online: https://www.nhs.uk/conditions/blood-groups/#:~:text=About%2085%25%20of%20the%20UK,type%20is%20not%20immediately%20known (accessed on 4 July 2022).
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33 (Suppl. 1), D514–D517. [Google Scholar] [CrossRef]
- Anstee, D.J. The relationship between blood groups and disease. J. Am. Soc. Hematol. 2010, 115, 4635–4643. [Google Scholar] [CrossRef] [PubMed]
- Simmons, R.T. Blood group genes in Polynesians and comparisons with other Pacific peoples. Oceania 1962, 32, 198–210. [Google Scholar] [CrossRef]
- Irshaid, N.M.; Henry, S.M.; Olsson, M.L. Genomic characterization of the Kidd blood group gene: Different molecular basis of the Jk (a–b–) phenotypein Polynesians and Finns. Transfusion 2000, 40, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, S.M.; Lenz, V.; Just, B.; Enczmann, J.; Fischer, J.C.; Horn, P.A.; Zeiler, T.A.; Balz, V. High-Throughput Next-Generation Sequencing of the Kidd Blood Group: Unexpected Antigen Expression Properties of Four Alleles and Detection of Novel Variants. Transfus. Med. Hemotherapy 2023, 50, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Seltsam, A.; Wagner, F.F.; Salama, A.; Flegel, W.A. Antibodies to high-frequency antigens may decrease the quality of transfusion support: An observational study. Transfusion 2003, 43, 1563–1566. [Google Scholar] [CrossRef] [PubMed]
- Scharberg, E.A.; Rink, G.; Schulz, D.; Rothenberger, S.; Stürtzel, A.; Gillhuber, N.; Seyboth, S.; Bugert, P. KDAS, a new blood group antigen in the Knops blood group system antithetical to KCAM. Transfusion 2020, 60, E25–E27. [Google Scholar] [CrossRef] [PubMed]
- Australian Red Cross Lifeblood. Major Blood Group Phenotypes and Their Frequencies. Blood Group Phenotypes 2021–2023. Available online: https://www.lifeblood.com.au/health-professionals/testing/blood-groups/phenotypes (accessed on 9 February 2023).
- Azouzi, S.; Mikdar, M.; Hermand, P.; Gautier, E.F.; Salnot, V.; Willemetz, A.; Nicolas, G.; Vrignaud, C.; Raneri, A.; Mayeux, P.; et al. Lack of the multidrug transporter MRP4/ABCC4 defines the PEL-negative blood group and impairs platelet aggregation. Blood. J. Am. Soc. Hematol. 2020, 135, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.E.; Lomas-Francis, C. The Blood Group Antigen Factsbook; Academic Press: Cambridge, MA, USA, 2012. [Google Scholar]
- McCarroll, S.A.; Kuruvilla, F.G.; Korn, J.M.; Cawley, S.; Nemesh, J.; Wysoker, A.; Shapero, M.H.; Bakker, P.I.W.d.; Maller, J.B.; Kirby, A.; et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008, 40, 1166–1174. [Google Scholar] [CrossRef]
- Mougey, R. A review of the Chido/Rodgers blood group. Immunohematology 2010, 26, 30–38. [Google Scholar] [CrossRef]
- Lundtoft, C.; Pucholt, P.; Martin, M.; Bianchi, M.; Lundström, E.; Eloranta, M.; Sandling, J.K.; Sjöwall, C.; Jönsen, A.; Gunnarsson, I.; et al. Complement C4 copy number variation is linked to SSA/Ro and SSB/La autoantibodies in systemic inflammatory autoimmune diseases. Arthritis Rheumatol. 2022, 74, 1440–1450. [Google Scholar] [CrossRef]
- Barber, M.R.; Drenkard, C.; Falasinnu, T.; Hoi, A.; Mak, A.; Kow, N.Y.; Svenungsson, E.; Peterson, J.; Clarke, A.E.; Ramsey-Goldman, R. Global epidemiology of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2021, 17, 515–532. [Google Scholar] [CrossRef]
- Gill, J.C.; Endres-Brooks, J.; Bauer, P.J.; Marks, W.J.; Montgomery, R.R. The effect of ABO blood group on the diagnosis of von Willebrand disease. Blood 1987, 69, 1691–1695. [Google Scholar] [CrossRef]

| Phenotype | Population | Frequency (%) | p-Value |
|---|---|---|---|
| D+ | NI | 96 (88.89) | - |
| AFR | 638 (96.52) | <0.0001 ** | |
| AMR | 326 (93.95) | 0.119 | |
| EAS | 503 (99.80) | <0.0001 ** | |
| EUR | 422 (83.90) | 0.245 | |
| SAS | 462 (94.48) | 0.056 | |
| D- | NI | 10 (9.26) | - |
| AFR | 23 (3.48) | 0.013 | |
| AMR | 21 (6.05) | 0.349 | |
| EAS | 1 (0.20) | NA | |
| EUR | 81 (16.10) | 0.096 | |
| SAS | 27 (5.52) | 0.216 | |
| Weak D (type 33) | NI | 2 (1.85) | - |
| AFR | 0 (0) | NA | |
| AMR | 0 (0) | NA | |
| EAS | 0 (0) | NA | |
| EUR | 0 (0) | NA | |
| SAS | 0 (0) | NA |
| Phenotype | Population | Frequency (%) | p-Value |
|---|---|---|---|
| Jk(a+b−) | NI | 37 (34.26) | - |
| AFR | 336 (50.83) | 0.001 | |
| AMR | 72 (20.75) | 0.006 | |
| EAS | 32 (6.35) | NA | |
| EUR | 124 (24.65) | 0.05 | |
| SAS | 155 (31.69) | 0.688 | |
| Jk(a+b+) | NI | 45 (41.67) | - |
| AFR | 126 (19.06) | <0.0001 ** | |
| AMR | 85 (24.49) | 0.001 | |
| EAS | 46 (9.13) | <0.0001 | |
| EUR | 169 (33.59) | 0.138 | |
| SAS | 99 (20.25) | <0.0001 ** | |
| Jk(a+b+w)/Jk(a+wb+) | NI | 10 (9.26) | - |
| AFR | 0 (0) | NA | |
| AMR | 0 (0) | NA | |
| EAS | 2 (0.4) | NA | |
| EUR | 0 (0) | NA | |
| SAS | 0 (0) | NA | |
| Jk(a−b+) | NI | 13 (12.04) | - |
| AFR | 55 (8.32) | 0.281 | |
| AMR | 102 (29.39) | <0.0001 ** | |
| EAS | 136 (26.98) | <0.0001 ** | |
| EUR | 129 (25.65) | 0.004 | |
| SAS | 68 (13.91) | 0.72 | |
| Jk(a+wb−) | NI | 3 (2.78) | - |
| AFR | 62 (38.58) | NA | |
| AMR | 22 (16.43) | NA | |
| EAS | 80 (21.43) | NA | |
| EUR | 19 (5.96) | NA | |
| SAS | 57 (28.22) | NA |
| Blood Group System | ISBT & Nucleotide Change | Phenotype | RBCeq Prediction | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Zygosity | NI% N = 108 | AFR% N = 661 | AMR% N = 347 | EAS% N = 504 | EUR% N = 503 | SAS% N = 489 | |||
| KN (ISBT 022) | KN*01.-08 c4828T>A | Kn(a+b−) McC(a+b−) Sl1+ Yk(a+) Vil- Sl3- KCAM+ | Heterozygous | 5.56 (n = 6) | 0.00 (n = 0) | 1.15 (n = 4) | 0.00 (n = 0) | 3.77 (n = 20) | 0.20 (n = 1) |
| KN (ISBT 022) | KN*01-05 c.4223C>T | Kn(a+b−) McC(a+b−) Sl1+ Yk(a−) Vil- Sl3+ KCAM+ | Homozygous | 13.89 (n = 15) | 0.30 (n = 2) | 16.14 (n = 56) | 12.70 (n = 64) | 9.94 (n = 49) | 10.84 (n = 53) |
| SC (ISBT 013) | SC*01.-07 | Sc1+ SCAN+ | Heterozygous | 2.78 (n = 3) | 0.00 (n = 0) | 0.86 (n = 3) | 0.00 (n = 0) | 0.99 (n = 5) | 0.20 (n = 1) |
| CO (ISBT 015) | CO*01/CO*02 c.134C>T | Co(a+b+) | Heterozygous | 6.48 (n = 7) | 1.82 (n = 12) | 1.73 (n = 6) | 0.00 (n = 0) | 6.56 (n = 33) | 0.82 (n = 4) |
| KEL (ISBT 006) | KEL*02 c.578C | K+k+ | Heterozygous | 3.70 (n = 4) | 0.45 (n = 3) | 4.32 (n = 15) | 0.00 (n = 0) | 7.55 (n = 38) | 1.23 (n = 6) |
| YT (ISBT 011) | YT*01/YT*02 c.1057C>A | Yt(a+b+) | Heterozygous | 6.48 (n = 7) | 0.00 (n = 0) | 7.20 (n = 25) | 0.40 (n = 2) | 9.54 (n = 48) | 8.38 (n = 41) |
| Blood Group System/Gene | Genomic Positions for Deletions | Impacted Intron/Exon | Frequency NI | Frequency Global | OMIM ID | Clinical Significance |
|---|---|---|---|---|---|---|
| CH/RG (ISBT 017) C4A | 31,995,858– 31,996,157 | Intron 25-Intron 26 | 4.63% | 6.51% | 120810 [30] | C4a deficiency [30] |
| 31,996,158– 31,996,457 | Intron 26-exon28 | 4.63% | 6.51% | 120810 [30] | C4a deficiency [30] | |
| 31,996,458– 31,996,757 | Exon 27-intron 28 | 5.55% | 6.51% | 120810 [30] | C4a deficiency [30] | |
| CH/RG (ISBT 017) C4B | 32,028,596– 32,028,895 | Intron 25-Intron 26 | 5.55% | 6.51% | 120820 [30] | C4B deficiency [30] |
| 32,028,896– 32,029,195 | Intron 26- Exon 28 | 7.40% | 6.51% | 120820 [30] | C4B deficiency [30] | |
| 32,029,196– 32,029,495 | Exon 28- Intron 28 | 7.40% | 6.51% | 120820 [30] | C4B deficiency [30] | |
| ABO (ISBT 001) | 133,255,603133,255,902 | Exon 7- Exon 7 | 1.85% | 1.29% | 616093 [30] | Blood group, ABO system [30] |
| Blood Group System | ISBT | Zygosity | RBCeq Prediction | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | NI% N = 108 | AFR% N = 661 | AMR% N = 347 | EAS% N = 504 | EUR% N = 503 | SAS% N = 489 | |||
| FY (ISBT 008) | FY*02W.01 | Heterozygous | Fy(a+b+w) OR Fy(a+wb+) OR Fyˣ | 0.93 (n = 1) | 0.00 (n = 0) | 0.00 (n = 0) | 0.00 (n = 0) | 0.00 (n = 0) | 0.00 (n = 0) |
| LW (ISBT 016) | LW*05/LW*07 | Heterozygous | Lw(a+b+) | 0.93 (n = 1) | 0 (n = 0) | 0 (n = 0) | 0 (n = 0) | 0.80 (n = 4) | 0 (n = 0) |
| KEL (ISBT 006) | KEL*02/KEL*02.03 | Heterozygous | K-k+ | 0.93 (n = 1) | 0.30 (n = 2) | 3.17 (n = 11) | 0.00 (n = 0) | 0.40 (n = 12) | 0.20 (n = 1) |
| PEL (ISBT 040) | ABCC4*01.02W c.912G>T | Homozygous | PEL+w | 0.93 (n = 1) | 5.3 (n = 35) | 0.29 (n = 1) | 3.17 (n = 16) | 0.99 (n = 5) | 2.66 (n = 13) |
| H (ISBT 018) | FUT2*01W.02.01 | Homozygous | H+w | 0.93 (n = 1) | 0.00 (n = 0) | 0.00 (n = 0) | 19.05 (n = 94) | 0.00 (n = 0) | 0.00 (n = 0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Brien, S.; Lea, R.A.; Jadhao, S.; Lee, S.; Sukhadia, S.; Arunachalam, V.; Roulis, E.; Flower, R.L.; Griffiths, L.; Nagaraj, S.H. Genetic Characterization of Blood Group Antigens for Polynesian Heritage Norfolk Island Residents. Genes 2023, 14, 1740. https://doi.org/10.3390/genes14091740
O’Brien S, Lea RA, Jadhao S, Lee S, Sukhadia S, Arunachalam V, Roulis E, Flower RL, Griffiths L, Nagaraj SH. Genetic Characterization of Blood Group Antigens for Polynesian Heritage Norfolk Island Residents. Genes. 2023; 14(9):1740. https://doi.org/10.3390/genes14091740
Chicago/Turabian StyleO’Brien, Stacie, Rodney A. Lea, Sudhir Jadhao, Simon Lee, Shrey Sukhadia, Vignesh Arunachalam, Eileen Roulis, Robert L. Flower, Lyn Griffiths, and Shivashankar H. Nagaraj. 2023. "Genetic Characterization of Blood Group Antigens for Polynesian Heritage Norfolk Island Residents" Genes 14, no. 9: 1740. https://doi.org/10.3390/genes14091740
APA StyleO’Brien, S., Lea, R. A., Jadhao, S., Lee, S., Sukhadia, S., Arunachalam, V., Roulis, E., Flower, R. L., Griffiths, L., & Nagaraj, S. H. (2023). Genetic Characterization of Blood Group Antigens for Polynesian Heritage Norfolk Island Residents. Genes, 14(9), 1740. https://doi.org/10.3390/genes14091740






