Genomic Complexity and Complex Chromosomal Rearrangements in Genetic Diagnosis: Two Illustrative Cases on Chromosome 7
Abstract
1. Introduction
2. Materials and Methods
2.1. Chromosome Analysis
2.2. FISH Analysis
2.3. Array Comparative Genomic Hybridization (Array-CGH)
2.4. In Silico Analysis
3. Results
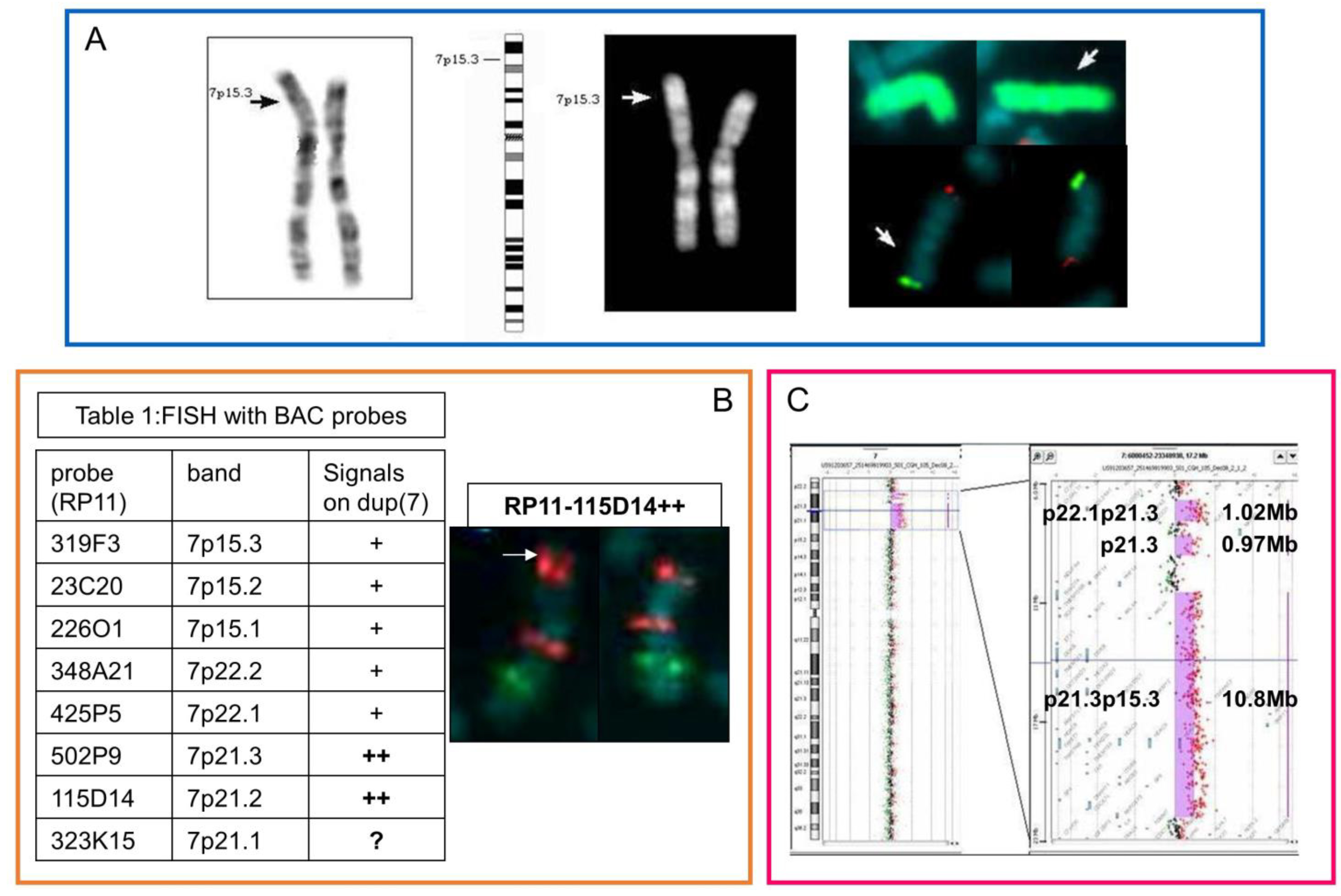
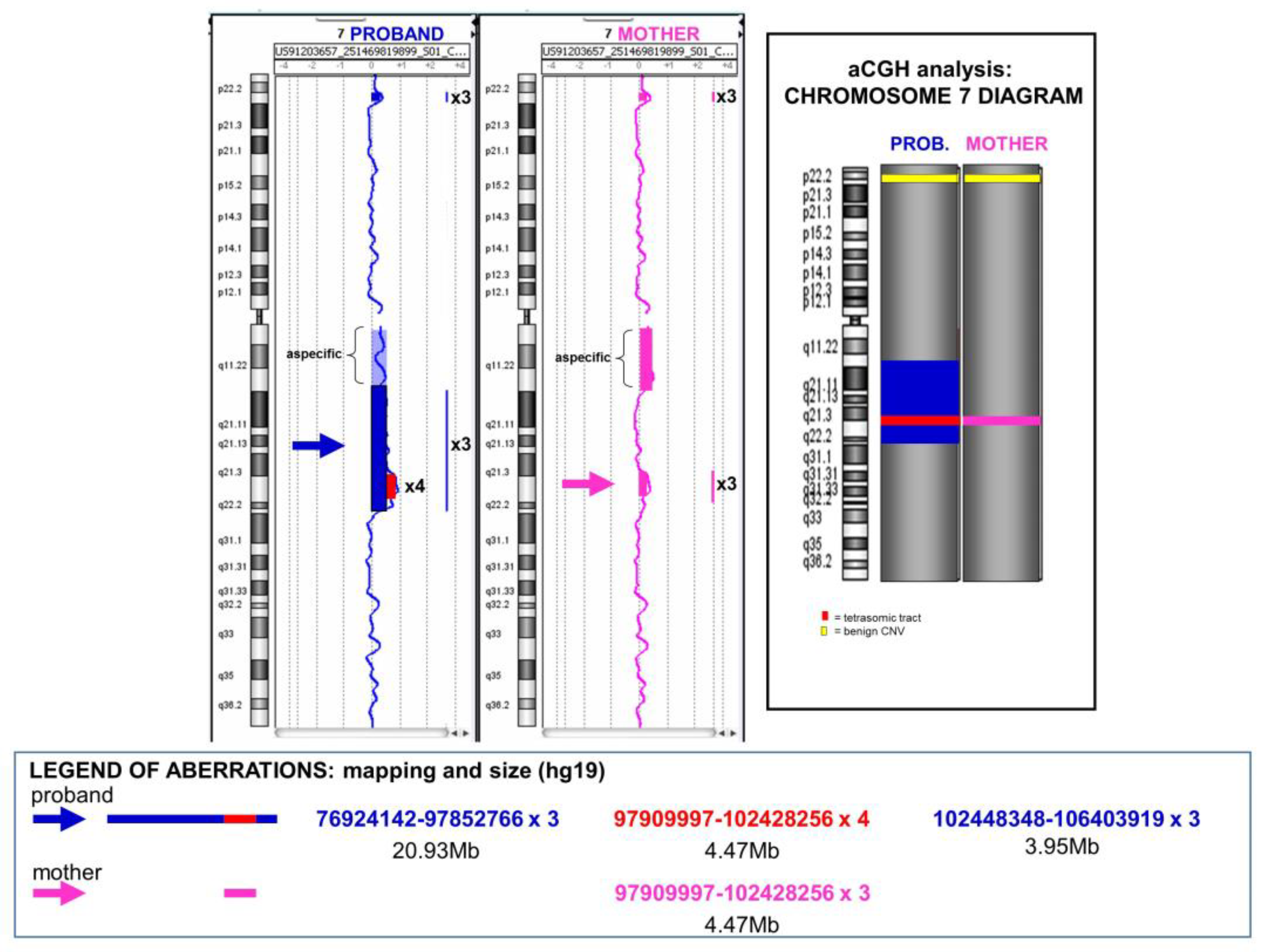
3.1. Case 1
3.1.1. Clinical Presentation
3.1.2. Laboratory Investigation
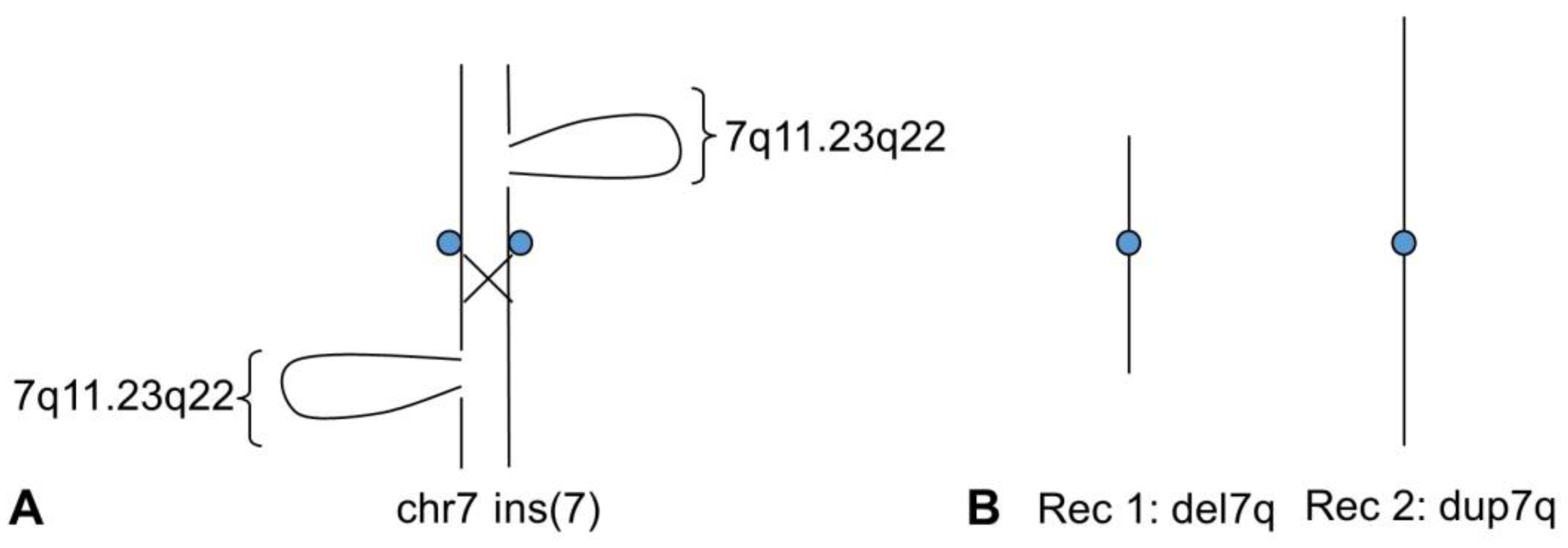
3.2. Case 2
3.2.1. Clinical Presentation
3.2.2. Laboratory Investigation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pellestor, F.; Anahory, T.; Lefort, G.; Puechberty, J.; Liehr, T.; Hedon, B.; Sarda, P. Complex chromosomal rearrangements: Origin and meiotic behavior. Hum. Reprod. Update 2011, 17, 476–494. [Google Scholar] [CrossRef]
- Poot, M.; Haaf, T. Mechanisms of Origin, Phenotypic Effects and Diagnostic Implications of Complex Chromosome Rearrangements. Mol. Syndromol. 2015, 6, 110–134. [Google Scholar] [CrossRef] [PubMed]
- Redaelli, S.; Conconi, D.; Sala, E.; Villa, N.; Crosti, F.; Roversi, G.; Bentivegna, A. Characterization of Chromosomal Breakpoints in 12 Cases with 8p Rearrangements Defines a Continuum of Fragility of the Region. Int. J. Mol. Sci. 2022, 23, 3347. [Google Scholar] [CrossRef] [PubMed]
- Michaelson-Cohen, R.; Murik, O.; Zeligson, S.; Lobel, O.; Weiss, O.; Picard, E.; Segel, R. Combining cytogenetic and genomic technologies for deciphering challenging complex chromosomal rearrangements. Mol. Genet. Genom. 2022, 297, 925–933. [Google Scholar] [CrossRef]
- Madan, K. What Is a Complex Chromosome Rearrangement? Am. J. Med. Genet. Part A 2013, 161A, 1181–1184. [Google Scholar] [CrossRef]
- Madan, K. Balanced complex chromosome rearrangements: Reproductive aspects. A review. Am. J. Med. Genet. A. 2012, 158A, 947–963. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zhang, F.; Lupski, J.R. Mechanisms for human genomic rearrangements. Pathogenetics 2008, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Lupski, J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002, 18, 74–82. [Google Scholar] [CrossRef]
- Liu, P.; Erez, A.; Nagamani, S.C.; Dhar, S.U.; Kołodziejska, K.E.; Dharmadhikari, A.V.; Cooper, M.L.; Wiszniewska, J.; Zhang, F.; Withers, M.A.; et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 2011, 146, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Pellestor, F.; Gatinois, V. Chromoanagenesis: A piece of the macroevolution scenario. Mol. Cytogenet. 2020, 13, 3. [Google Scholar] [CrossRef]
- McGowan-Jordan, J.; Hastings, R.J.; Moore, S. An International System for Human Cytogenomic Nomenclature; Karger: Basel, Switzerland, 2020. [Google Scholar]
- Carvalho, C.M.; Zhang, F.; Liu, P.; Patel, A.; Sahoo, T.; Bacino, C.A.; Shaw, C.; Peacock, S.; Pursley, A.; Tavyev, Y.J.; et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum. Mol. Genet. 2009, 18, 2188–2203. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Shaw, C.J.; Dapper, J.D.; Wakui, K.; Shaffer, L.G.; Withers, M.; Lupski, J.R. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am. J. Hum. Genet. 2003, 72, 1101–1116. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hillier, L.W.; Fulton, R.S.; Fulton, L.A.; Graves, T.A.; Pepin, K.H.; Wagner-McPherson, C.; Wilson, R.K. The DNA sequence of human chromosome 7. Nature 2003, 424, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Horvath, J.E.; Bailey, J.A.; Locke, D.P.; Eichler, E.E. Lessons from the human genome: Transitions between euchromatin and heterochromatin. Hum. Mol. Genet. 2001, 10, 2215–2223. [Google Scholar] [CrossRef]
- Mefford, H.C.; Trask, B.J. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 2002, 3, 91–102. [Google Scholar] [CrossRef]
- Stankiewicz, P.; Lupski, J.R. Molecular-evolutionary mechanisms for genomic disorders. Curr. Opin. Genet. Dev. 2002, 12, 312–319. [Google Scholar] [CrossRef]
- Lee, J.A.; Carvalho, C.M.; Lupski, J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 2007, 131, 1235–1247. [Google Scholar] [CrossRef]
- Hastings, P.J.; Ira, G.; Lupski, J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009, 5, e1000327. [Google Scholar] [CrossRef]
- AlFardan, J.; Brown, K.; Gessner, J.; Lunt, B.; Scharer, G. Small duplication of chromosome (7)(p22.1p22.2) and consideration of a dup 7p syndrome critical region. Clin. Dysmorphol. 2011, 20, 217–221. [Google Scholar] [CrossRef]
- Stankiewicz, P.; Thiele, H.; Baldermann, C.; Krüger, A.; Giannakudis, I.; Dörr, S.; Werner, N.; Kunz, J.; Rappold, G.A.; Hansmann, I. Phenotypic findings due to trisomy 7p15.3-pter including the TWIST locus. Am. J. Med. Genet. 2001, 103, 56–62. [Google Scholar] [CrossRef]
- Mégarbané, A.; Le Lorc’H, M.; Elghezal, H.; Joly, G.; Gosset, P.; Souraty, N.; Romana, S.P. Pure partial 7p trisomy including the TWIST, HOXA, and GLI3 genes. J. Med. Genet. 2001, 38, 178–182. [Google Scholar] [CrossRef][Green Version]
- Romanelli Tavares, V.L.; Guimarães-Ramos, S.L.; Zhou, Y.; Masotti, C.; Ezquina, S.; de Paula Moreira, D.; Passos-Bueno, M.R. New locus underlying auriculocondylar syndrome (ARCND): 430 kb duplication involving TWIST1 regulatory elements. J. Med. Genet. 2022, 59, 895–905. [Google Scholar] [PubMed]
- Schwenk, B.M.; Lang, C.M.; Hogl, S.; Tahirovic, S.; Orozco, D.; Rentzsch, K.; Lichtenthaler, S.F.; Hoogenraad, C.C.; Capell, A.; Haass, C.; et al. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J. 2014, 3, 450–467. [Google Scholar] [CrossRef] [PubMed]
- Brady, O.A.; Zheng, Y.; Murphy, K.; Huang, M.; Hu, F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet. 2013, 22, 685–695. [Google Scholar] [CrossRef]
- Alfonsi, M.; Palka, C.; Morizio, E.; Gatta, V.; Franchi, S.; Franchi, P.G.; Zori, R.; Calabrese, G.; Palka, G.; Chiarelli, F. A new case of pure partial 7q duplication. Cytogenet. Genome Res. 2012, 136, 1–5. [Google Scholar] [CrossRef]
- Tchirikov, M.; Merinsky, A.A.; Strohner, M.; Bonin, M.; Beyer, V.; Haaf, T.; Bartsch, O. Prenatal diagnosis of a recombinant chromosome 7 resulting in trisomy 7q11.22 --> qter. Am. J. Med. Genet. A 2010, 152A, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Scelsa, B.; Bedeschi, F.M.; Guerneri, S.; Lalatta, F.; Introvini, P. Partial Trisomy of 7q: Case Report and Literature Review. J. Child Neurol. 2008, 23, 572–579. [Google Scholar] [PubMed]
- Siewert, S.E.; Della Vedova, M.C.; Guillamondegui, M.J.; Drut, M.; Brezigar, A.; Cardetti, M.; Marsá, S.M. A newborn with partial pure trisomy of chromosome 7q inherited from paternal balanced translocation with congenital anomalies. a mini review. Indian J. Med. Res. Pharm. Sci. 2018, 5, 82–91. [Google Scholar]
- Ishii, F.; Fujita, H.; Nagai, A.; Ogihara, T.; Kim, H.-S.; Okamoto, R.; Mino, M. Case report of rec(7)dup(7q)inv(7)(p22q22) and a review of the recombinants resulting from parental pericentric inversions on any chromosomes. Am. J. Med. Genet. 1997, 73, 290–295. [Google Scholar] [CrossRef]
- Catusi, I.; Bonati, M.T.; Mainini, E.; Russo, S.; Orlandini, E.; Larizza, L.; Recalcati, M.P. Recombinant Chromosome 7 Driven by Maternal Chromosome 7 Pericentric Inversion in a Girl with Features of Silver-Russell Syndrome. Int. J. Mol. Sci. 2020, 21, 8487. [Google Scholar] [CrossRef]
- Udayakumar, A.M.; Al-Mamari, W.; Al-Sayegh, A.; Al-Kindy, A. De Novo Duplication of 7p21.1p22.2 in a Child with Autism Spectrum Disorder and Craniofacial Dysmorphism. Sultan Qaboos Univ. Med. J. 2015, 15, e415–e419. [Google Scholar] [CrossRef] [PubMed]

| Case | Case 1 | Case 2 |
|---|---|---|
| Age at diagnosis (Days, Months, Years) | 28 m | 5 d |
| Sex (M/F) | F | M |
| Major Malformation | ||
| Criptorchid testes | + | |
| Heart defect | − | |
| Renal abnormalities | − | + |
| Cerebral Abnormalities (MRI) | − | |
| Minor Anomalies | ||
| Prominent/high forehead | − | |
| Hypertelorism | − | |
| Broad and/or flat nasal bridge | + | |
| High palate | + | |
| Thin lip | + | |
| Micrognathia | + | |
| Abnormal palmar creases | + | |
| Skeletal anomalies | ||
| Short neck | + | |
| Digital anomalies | + | |
| Medical Complications | ||
| Hypotonia | + | |
| Ataxia | + | |
| Epilepsy | + | |
| Growth and Development | ||
| Small at birth | − | |
| Developmental delay | + | |
| Stereotypic behavior | + | |
| Mental retardation | + | |
| Language impairment | + | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villa, N.; Redaelli, S.; Farina, S.; Conconi, D.; Sala, E.M.; Crosti, F.; Mariani, S.; Colombo, C.M.; Dalprà, L.; Lavitrano, M.; et al. Genomic Complexity and Complex Chromosomal Rearrangements in Genetic Diagnosis: Two Illustrative Cases on Chromosome 7. Genes 2023, 14, 1700. https://doi.org/10.3390/genes14091700
Villa N, Redaelli S, Farina S, Conconi D, Sala EM, Crosti F, Mariani S, Colombo CM, Dalprà L, Lavitrano M, et al. Genomic Complexity and Complex Chromosomal Rearrangements in Genetic Diagnosis: Two Illustrative Cases on Chromosome 7. Genes. 2023; 14(9):1700. https://doi.org/10.3390/genes14091700
Chicago/Turabian StyleVilla, Nicoletta, Serena Redaelli, Stefania Farina, Donatella Conconi, Elena Maria Sala, Francesca Crosti, Silvana Mariani, Carla Maria Colombo, Leda Dalprà, Marialuisa Lavitrano, and et al. 2023. "Genomic Complexity and Complex Chromosomal Rearrangements in Genetic Diagnosis: Two Illustrative Cases on Chromosome 7" Genes 14, no. 9: 1700. https://doi.org/10.3390/genes14091700
APA StyleVilla, N., Redaelli, S., Farina, S., Conconi, D., Sala, E. M., Crosti, F., Mariani, S., Colombo, C. M., Dalprà, L., Lavitrano, M., Bentivegna, A., & Roversi, G. (2023). Genomic Complexity and Complex Chromosomal Rearrangements in Genetic Diagnosis: Two Illustrative Cases on Chromosome 7. Genes, 14(9), 1700. https://doi.org/10.3390/genes14091700







