MAGI-ACMG: Algorithm for the Classification of Variants According to ACMG and ACGS Recommendations
Abstract
1. Introduction
2. Materials and Methods
2.1. MAGI-ACMG Algorithm Description
- PM2: if the criterion is triggered at a Supporting or Strong level, this algorithm re-assigns the standard Moderate intensity;
- BP6: if the criterion is triggered at a Very Strong or Moderate level, the algorithm re-assigns a Supporting intensity. A subsequent strength confirmation is afterwards performed by the geneticist to confirm or upgrade the criteria to a Strong level.
- PP3:
- -
- For missense variants, at least 2/3 of the following must be true: (1) REVEL score ≥ 0.644; (2) CADD (score ≥ 25.3); (3) 8/15 of the rank scores of the 15 funtional predictors (DANN, EigenPC, FATHMM, LRT, MCAP, MetaLR, MetaSVM, MutPred, MutationAssessor, MutationTaster, PROVEAN, Polyphen2HDIV, Polyphen2HVAR, SIFT, VEST3) ≥ 0.644;
- -
- For missense and splicing variants: calculated as for missense variants, if not applicable assigned if AdaBoost score ≥ 0.708 AND RF score ≥ 0.515;
- -
- For synonymous and splicing variants: assigned if AdaBoost score ≥ 0.708 AND RF score ≥ 0.515;
- -
- For intronic variants (±5): assigned if AdaBoost score ≥ 0.708 AND RF score ≥ 0.515.
- BP4:
- -
- For missense variants, at least 2/3 of the following: REVEL score ≤ 0.29; CADD score ≤ 22.7; 8/15 of the rank scores of the 15 funtional predictors (DANN, EigenPC, FATHMM, LRT, MCAP, MetaLR, MetaSVM, MutPred, MutationAssessor, MutationTaster, PROVEAN, Polyphen2HDIV, Polyphen2HVAR, SIFT, VEST3) ≤ 0.29;
- -
- For missense and splicing variants: calculated as for missense variants, if not applicable it is assigned if AdaBoost score ≤ 0.708 and RF score ≤ 0.515;
- -
- Intronic and splicing (a ± 5): assigned if AdaBoost score ≤ 0.708 and RF score ≤ 0.515.
2.2. Subclassification of Variants of Uncertain Significance
- If in the same individual another pathogenic/likely pathogenic/Hot/Middle VUS is identified in a gene associated with an autosomal recessive disorder;
- If the middle VUS is in the homozygous state in a gene associated with autosomal recessive disorder;
- If the middle VUS is in the heterozygous state in a gene associated with an autosomal dominant disorder;
- If the middle VUS is the hemizygous state in a gene associated with an X-linked condition.
- 1 PM criterion + 2 PP criteria;
- 2 PM criteria.
2.3. Implementation of MAGI ACMG Web Application
3. Results
3.1. In Silico Predictors, Application of Criteria BP4/PP3
3.2. AutoPVS1 Implementation
3.3. Segregation Analysis: P_POT (for In-Trans Allelic Configuration)
3.4. Segregation Analysis: P_POT (for De Novo Occurrence)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Abou Tayoun, A.N.; Pesaran, T.; DiStefano, M.T.; Oza, A.; Rehm, H.L.; Biesecker, L.G.; Harrison, S.M. Recommendations for Interpreting the Loss of Function PVS1 ACMG/AMP Variant Criterion. Hum. Mutat. 2018, 39, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Brnich, S.E.; Abou Tayoun, A.N.; Couch, F.J.; Cutting, G.R.; Greenblatt, M.S.; Heinen, C.D.; Kanavy, D.M.; Luo, X.; McNulty, S.M.; Starita, L.M.; et al. Recommendations for Application of the Functional Evidence PS3/BS3 Criterion Using the ACMG/AMP Sequence Variant Interpretation Framework. Genome Med. 2020, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Byrne, A.B.; Feng, B.-J.; Pagel, K.A.; Mooney, S.D.; Karchin, R.; O’Donnell-Luria, A.; Harrison, S.M.; Tavtigian, S.V.; Greenblatt, M.S.; et al. Calibration of Computational Tools for Missense Variant Pathogenicity Classification and ClinGen Recommendations for PP3/BP4 Criteria. Am. J. Hum. Genet. 2022, 109, 2163–2177. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.A.; Caleshu, C.; Morales, A.; Buchan, J.; Wolf, Z.; Harrison, S.M.; Cook, S.; Dillon, M.W.; Garcia, J.; Haverfield, E.; et al. Adaptation and Validation of the ACMG/AMP Variant Classification Framework for MYH7-Associated Inherited Cardiomyopathies: Recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet. Med. 2018, 20, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Gelb, B.D.; Cavé, H.; Dillon, M.W.; Gripp, K.W.; Lee, J.A.; Mason-Suares, H.; Rauen, K.A.; Williams, B.; Zenker, M.; Vincent, L.M. ClinGen’s RASopathy Expert Panel Consensus Methods for Variant Interpretation. Genet. Med. 2018, 20, 1334–1345. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The Human Genomic Variant Search Engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017, 100, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Preston, C.G.; Wright, M.W.; Madhavrao, R.; Harrison, S.M.; Goldstein, J.L.; Luo, X.; Wand, H.; Wulf, B.; Cheung, G.; Mandell, M.E.; et al. ClinGen Variant Curation Interface: A Variant Classification Platform for the Application of Evidence Criteria from ACMG/AMP Guidelines. Genome Med. 2022, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, E.; Cristofoli, F.; Modena, C.; Paolacci, S.; Bertelli, M.; Marceddu, G. Integration of VarSome API in an Existing Bioinformatic Pipeline for Automated ACMG Interpretation of Clinical Variants. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1–6. [Google Scholar] [PubMed]
- Cristofoli, F.; Sorrentino, E.; Guerri, G.; Miotto, R.; Romanelli, R.; Zulian, A.; Cecchin, S.; Paolacci, S.; Miertus, J.; Bertelli, M.; et al. Variant Selection and Interpretation: An Example of Modified VarSome Classifier of ACMG Guidelines in the Diagnostic Setting. Genes 2021, 12, 1885. [Google Scholar] [CrossRef] [PubMed]
- Ellard, S.; Baple, E.L.; Callaway, A.; Berry, I.; Forrester, N.; Turnbull, C.; Owens, M.; Eccles, D.M.; Abbs, S.; Scott, R.; et al. ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2020. 2020. Available online: https://www.acgs.uk.Com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf (accessed on 1 June 2023).
- Xiang, J.; Peng, J.; Baxter, S.; Peng, Z. AutoPVS1: An Automatic Classification Tool for PVS1 Interpretation of Null Variants. Hum. Mutat. 2020, 41, 1488–1498. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. DbNSFP v4: A Comprehensive Database of Transcript-Specific Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Jian, X.; Boerwinkle, E.; Liu, X. In Silico Tools for Splicing Defect Prediction: A Survey from the Viewpoint of End Users. Genet. Med. 2014, 16, 497–503. [Google Scholar] [CrossRef]

| VUS Class | Criteria Combination | ||||
|---|---|---|---|---|---|
| HOT | PVS | ||||
| BP | PVS | ||||
| PP | PVS | ||||
| PS | |||||
| PP | PS | ||||
| BP | PP | PS | |||
| BS | PP | PS | |||
| BP | PS | ||||
| PP | PM | PM | |||
| BP | PP | PM | PM | ||
| BS | PP | PM | PM | ||
| PP | PP | PP | PM | ||
| BP | PP | PP | PP | PM | |
| BS | PP | PP | PP | PM | |
| PP | PP | PP | PP | ||
| BP | PP | PP | PP | PP | |
| MIDDLE | PM | PM | |||
| BP | PM | PM | |||
| PP | PM | ||||
| BP | PP | PM | |||
| PP | PP | PM | |||
| BP | PP | PP | PM | ||
| BS | PVS | ||||
| BS | PS | ||||
| PP | PP | PP | |||
| BP | PP | PP | PP | ||
| COLD | PM | ||||
| BP | PM | ||||
| PP | PP | ||||
| BP | PP | PP | |||
| BS | PP | PP | |||
| PP | |||||
| BP | PP | ||||
| BS | PP | ||||
| BP | |||||
| BS | |||||
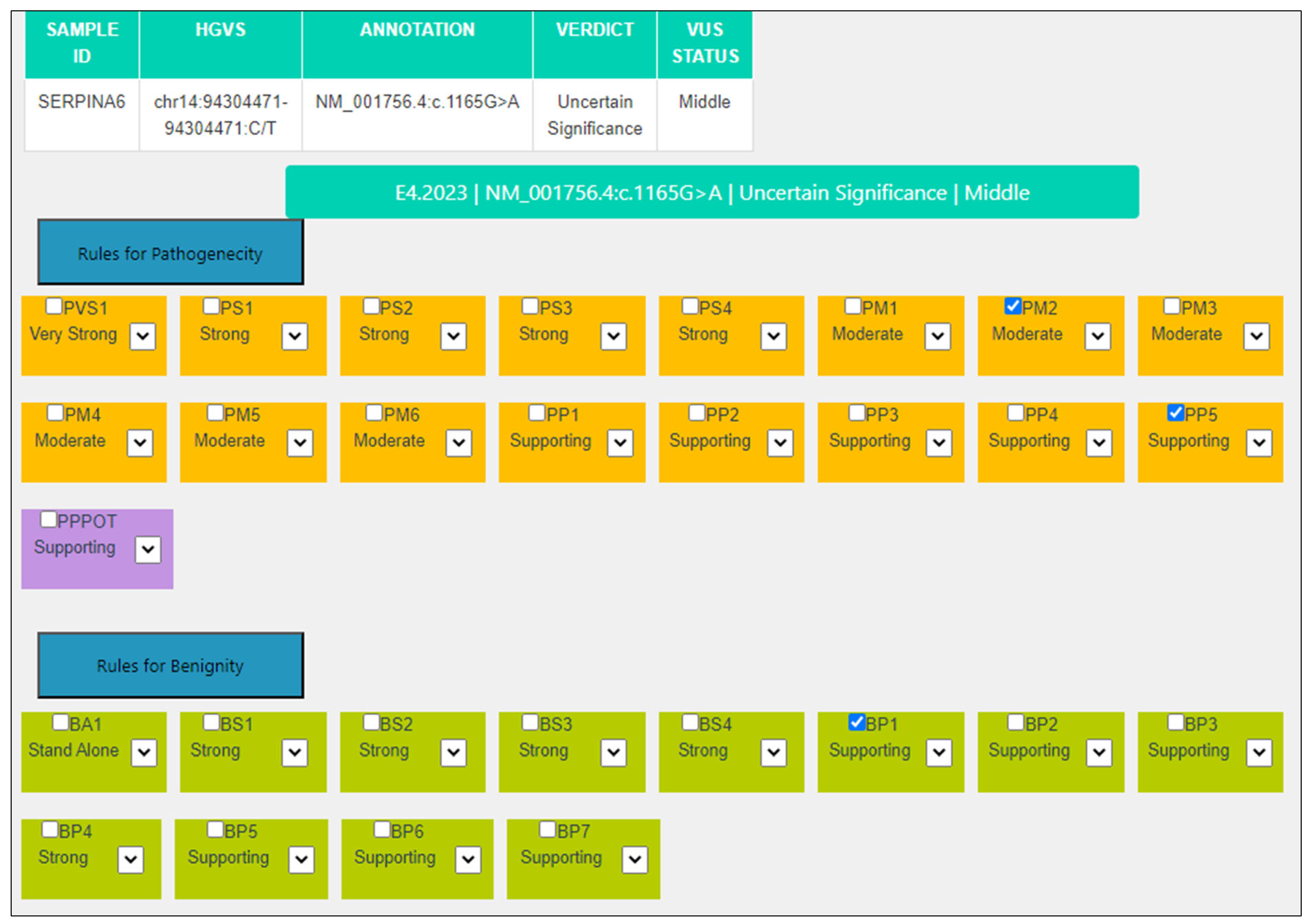
| Criterion | VarSome Stable-API v.11.6.1 | MAGI-ACMG |
|---|---|---|
| BP4 | Strong: MetaRNN = 0.0134 is between 0.00692 and 0.108. | Not applicable: 11/15 predictors are damaging, REVEL score: 0.518 (>0.29) and CADD score is 24.78 (>22.7). |
| PP3 | Not applicable | Not applicable: 11/15 predictors are damaging but REVEL score: 0.518 (<0.644) and CADD score is 24.78 (<25.3). |
| BP1 | Supporting: GnomAD missense Z-score for gene SERPINA6 is −0.662 which is less than 2.99. | Supporting: GnomAD missense Z-score for gene SERPINA6 is -0.662, which is less than 2.99. |
| PM2 | Supporting: GnomAD genomes homozygous allele count = 0 is less than 2 for AD/AR gene SERPINA6, good gnomAD genomes coverage = 30.9. | Moderate: Absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium. |
| PP5 | Supporting: ClinVar classifies this variant as Uncertain Significance, 2 stars (multiple consistent, reviewed Apr ‘22, 5 submissions), citing 3 articles (PUBMED:17245537, PUBMED:12780753, PUBMED:10634411), associated with Corticosteroid-Binding Globulin Deficiency, with 5 submissions (1 P, 2 LP and 2 VUS). | Supporting: ClinVar classifies this variant as Uncertain. Significance, 2 stars (multiple consistent, reviewed Apr ‘22, 5 submissions), citing 3 articles (PUBMED:17245537, PUBMED:12780753, PUBMED:10634411), associated with Corticosteroid-Binding Globulin Deficiency, with 5 submissions (1 P, 2 LP and 2 VUS). |
| Final | VUS | VUS Middle |
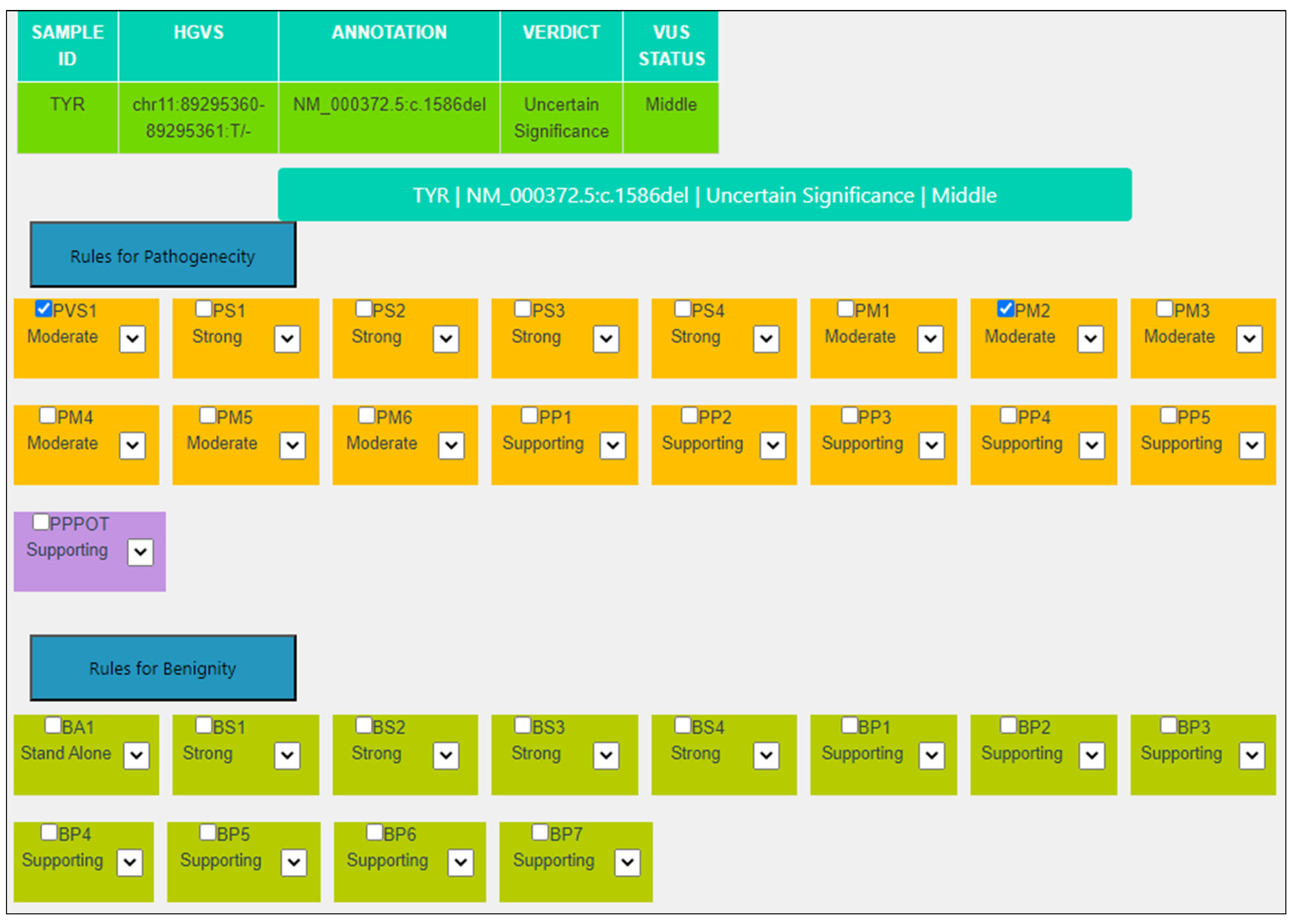
| Criterion | VarSome Stable-API v.11.6.1 | MAGI-ACMG |
|---|---|---|
| PVS1 | Strong: Null variant (frame-shift) in gene TYR, not predicted to cause NMD. Loss-of-function is a known mechanism of disease (gene has 78 reported pathogenic LOF variants). The truncated region contains 0 pathogenic variants. It removes 0.19% of the protein. | Moderate: NP6—LoF variants in this exon are not frequent in the general population and exon is present in biologically relevant transcript(s)—Variants remove <10% of protein. |
| PM2 | Supporting: Variant not found in gnomAD genomes, good gnomAD genomes coverage = 31.3. | Moderate: Absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium. |
| Final | VUS | VUS Middle |
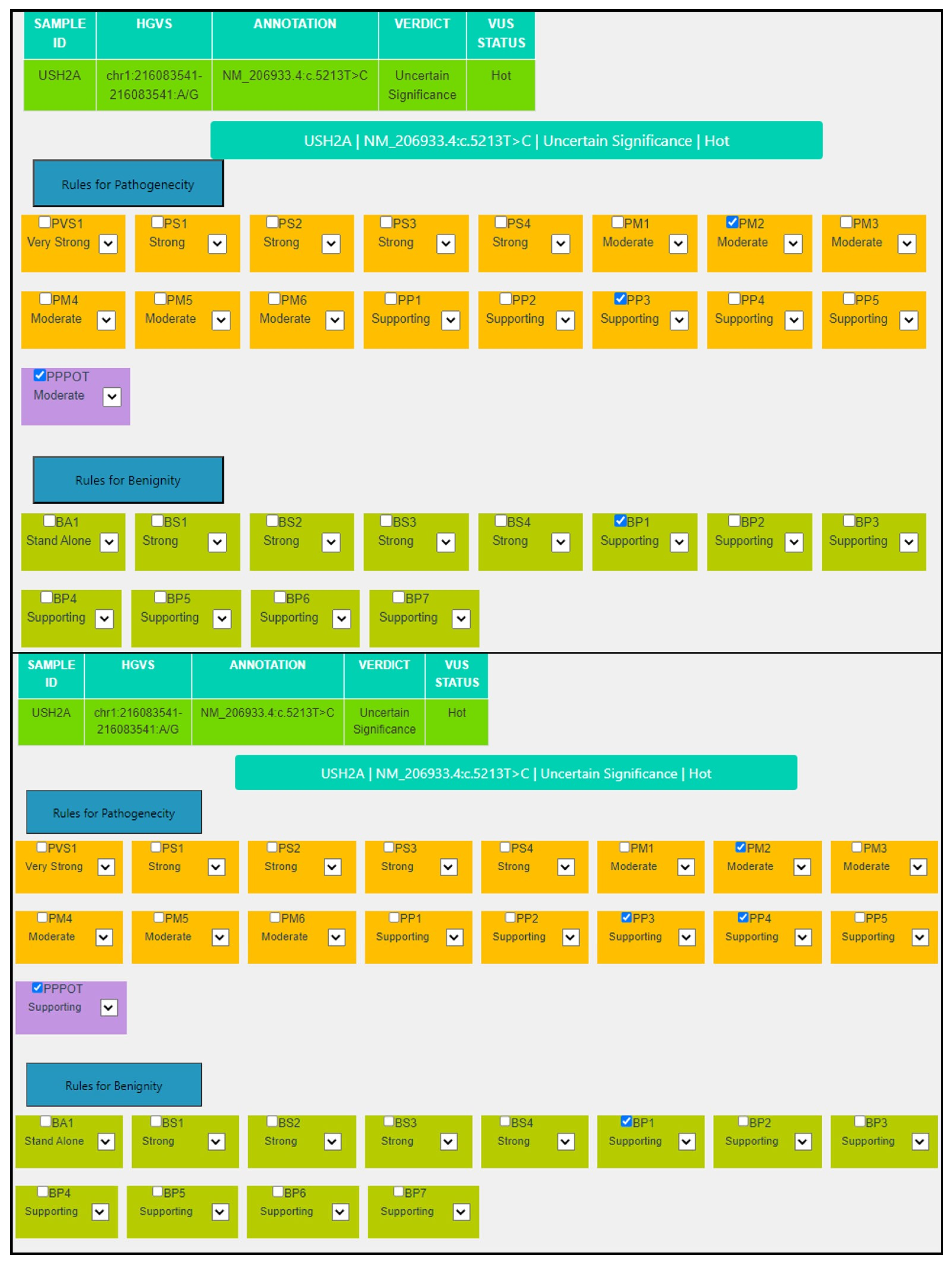
| Criterion | VarSome Stable-API v.11.6.1 | MAGI-ACMG |
|---|---|---|
| PP3 | Moderate: MetaRNN = 0.859 is between 0.841 and 0.939, moderate pathogenic. | Supporting: 14/15 predictors are damaging, REVEL SCORE: 0.767 (>0.644) and CADD score: 28.30 (>25.3). |
| BP1 | Supporting: 196 out of 529 non-VUS missense variants in gene USH2A are benign = 37.1% which is more than threshold of 33.1%. | Supporting: 196 out of 529 non-VUS missense variants in gene USH2A are benign = 37.1%, which is more than threshold of 33.1%. |
| PM2 | Supporting: Variant not found in gnomAD genomes, good gnomAD genomes coverage = 31.0. | Moderate: Absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium. |
| PP4 | Not applicable (case specific). | Applied |
| P-Pot | Supporting: possible in-trans configuration with the pathogenic USH2A:NM_206933.4:c.2299del variant. | |
| Final | VUS | VUS Hot |
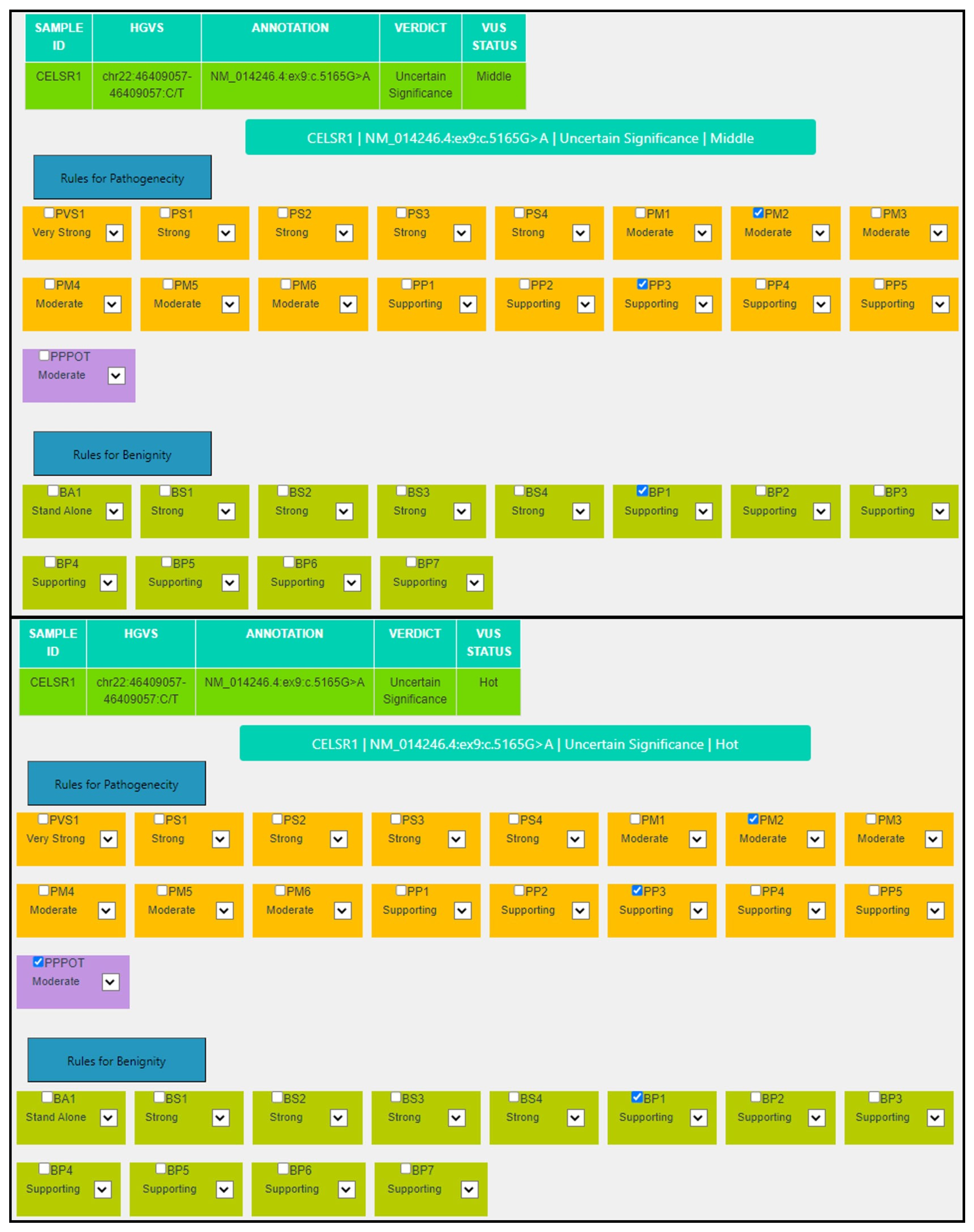
| Criterion | VarSome Stable-API v.11.6.1 | MAGI-ACMG |
|---|---|---|
| PP3 | Moderate: MetaRNN = 0.92 is between 0.841 and 0.939 ⇒ moderate pathogenic. | Supporting: 13/15 predictors are damaging, Revel score: 0.723 (>0.644) and CADD score: 29.1 (>25.3). |
| BP1 | Supporting: 33 out of 33 non-VUS missense variants in gene CELSR1 are benign = 100.0% which is more than threshold of 33.1%. | Supporting: 33 out of 33 non-VUS missense variants in gene CELSR1 are benign = 100.0%, which is more than threshold of 33.1%. |
| PM2 | Supporting: GnomAD genomes allele count = 2 is less than 5 for AD gene CELSR1, good gnomAD genomes coverage = 31.9 | Moderate: Absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium. |
| P-Pot | Moderate: possible de novo variant. | |
| Final | VUS | VUS Hot |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cristofoli, F.; Daja, M.; Maltese, P.E.; Guerri, G.; Tanzi, B.; Miotto, R.; Bonetti, G.; Miertus, J.; Chiurazzi, P.; Stuppia, L.; et al. MAGI-ACMG: Algorithm for the Classification of Variants According to ACMG and ACGS Recommendations. Genes 2023, 14, 1600. https://doi.org/10.3390/genes14081600
Cristofoli F, Daja M, Maltese PE, Guerri G, Tanzi B, Miotto R, Bonetti G, Miertus J, Chiurazzi P, Stuppia L, et al. MAGI-ACMG: Algorithm for the Classification of Variants According to ACMG and ACGS Recommendations. Genes. 2023; 14(8):1600. https://doi.org/10.3390/genes14081600
Chicago/Turabian StyleCristofoli, Francesca, Muharrem Daja, Paolo Enrico Maltese, Giulia Guerri, Benedetta Tanzi, Roberta Miotto, Gabriele Bonetti, Jan Miertus, Pietro Chiurazzi, Liborio Stuppia, and et al. 2023. "MAGI-ACMG: Algorithm for the Classification of Variants According to ACMG and ACGS Recommendations" Genes 14, no. 8: 1600. https://doi.org/10.3390/genes14081600
APA StyleCristofoli, F., Daja, M., Maltese, P. E., Guerri, G., Tanzi, B., Miotto, R., Bonetti, G., Miertus, J., Chiurazzi, P., Stuppia, L., Gatta, V., Cecchin, S., Bertelli, M., & Marceddu, G. (2023). MAGI-ACMG: Algorithm for the Classification of Variants According to ACMG and ACGS Recommendations. Genes, 14(8), 1600. https://doi.org/10.3390/genes14081600








