QTL Mapping of Tiller Number in Korean Japonica Rice Varieties
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth and DNA Extraction
2.2. Phenotypic Evaluations
2.3. Mapping and Identification of QTL
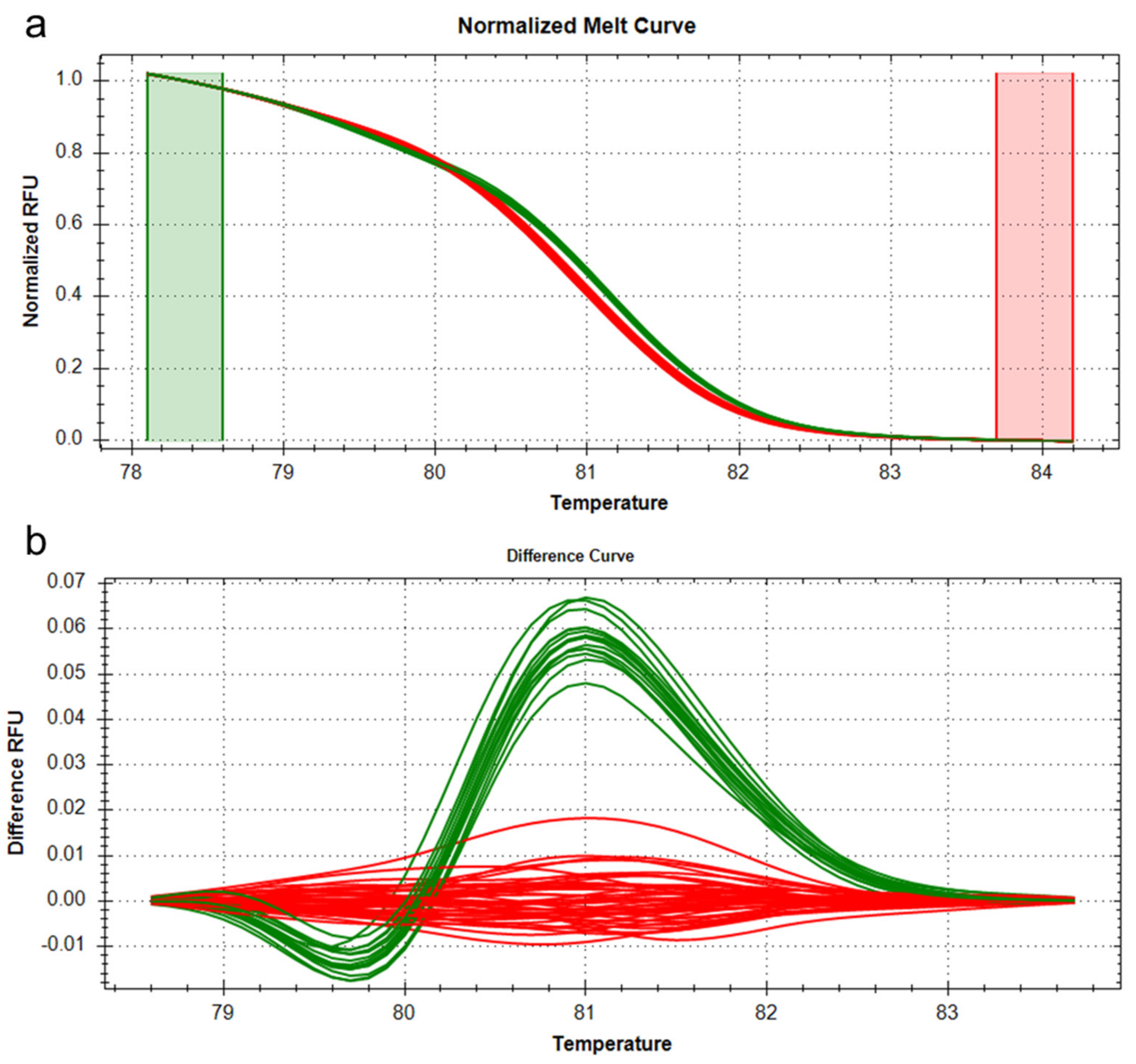
2.4. Analysis of OsTB1, a Candidate Gene for qTN3 on Chromosome 3
3. Results
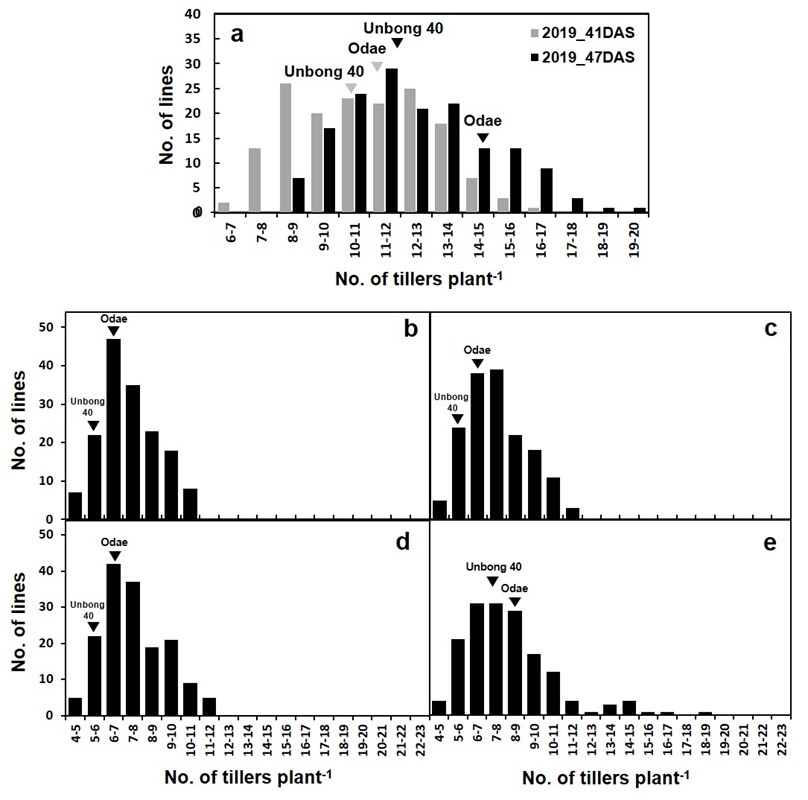
3.1. Analysis of Phenotypic Variation
3.2. QTL Mapping of Tiller Number
3.3. QTL Mapping for Some other Yield-Related Traits
3.4. Analysis of Candidate Genes for the QTL qTN3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data (accessed on 10 January 2023).
- Makino, A.; Kaneta, Y.; Obara, M.; Ishiyama, K.; Kanno, K.; Kondo, E.; Suzuki, Y.; Mae, T. High yielding ability of a large-grain rice cultivar, Akita 63. Sci. Rep. 2020, 10, 12231. [Google Scholar] [CrossRef]
- Hardy, B.; Sheehy, J.E.; Mitchell, P.L. Redesigning Rice Photosynthesis to Increase Yield; Elsevier Science: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Khush, G.S.; Peng, S. Improving Yield Potential by Modifying Plant Type; International Rice Research Institute (IRRI): Los Baños, Philippines, 1998. [Google Scholar] [CrossRef]
- Xu, Y.B.; Shen, Z.T. Diallel analysis of tiller number at different growth stages in rice (Oryza sativa L.). Theor. Appl. Genet. 1991, 83, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Zheng, H.L.; Wang, J.G.; Liu, H.L.; Sun, J.; Zhao, H.W.; Yang, L.M.; Zou, D.T. Genetic dissection of rice (Oryza sativa L.) tiller, plant height, and grain yield based on QTL mapping and metaanalysis. Euphytica 2018, 214, 109. [Google Scholar] [CrossRef]
- Zhao, D.-D.; Park, J.-R.; Jang, Y.-H.; Kim, E.-G.; Du, X.-X.; Farooq, M.; Yun, B.-J.; Kim, K.-M. Identification of One Major QTL and a Novel Gene OsIAA17q5 Associated with Tiller Number in Rice Using QTL Analysis. Plants 2022, 11, 538. [Google Scholar] [CrossRef]
- Kelly, S.A.; Panhuis, T.M.; Stoehr, A.M. Phenotypic plasticity: Molecular mechanisms and adaptive significance. Compr. Physiol. 2012, 2, 1417–1439. [Google Scholar] [CrossRef]
- Hussien, A.; Tavakol, E.; Horner, D.S.; Muñoz-Amatriaín, M.; Muehlbauer, G.J.; Rossini, L. Genetics of Tillering in Rice and Barley. Plant Genome 2014, 7. [Google Scholar] [CrossRef]
- Fujita, D.; Ebron, L.A.; Araki, E.; Kato, H.; Khush, G.S.; Sheehy, J.E.; Lafarge, T.; Fukuta, Y.; Kobayashi, N. Fine mapping of a gene for low-tiller number, Ltn, in japonica rice (Oryza sativa L.) variety Aikawa 1. Theor. Appl. Genet. 2010, 120, 1233–1240. [Google Scholar] [CrossRef]
- Sakamoto, T.; Matsuoka, M. Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 2004, 15, 144–147. [Google Scholar] [CrossRef]
- MÄKelÄ, P.; Muurinen, S. Uniculm and conventional tillering barley accessions under northern growing conditions. J. Agric. Sci. 2011, 150, 335–344. [Google Scholar] [CrossRef]
- Takai, T.; Fukuta, Y.; Shiraiwa, T.; Horie, T. Time-related mapping of quantitative trait loci controlling grain-filling in rice (Oryza sativa L.). J. Exp. Bot. 2005, 56, 2107–2118. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Zhu, G.; Zhu, C.; Peng, X.; Li, C.; He, X.; Chen, X.; Fu, J.; Hu, L.; Ouyang, L.; et al. Molecular dissection of developmental behavior of tiller number and the relationship with effective panicle using indica–japonica introgression lines in rice. Mol. Breed. 2015, 35, 91. [Google Scholar] [CrossRef]
- Kubo, T.; Aida, Y.; Nakamura, K.; Tsunematsu, H.; Doi, K.; Yoshimura, A. Reciprocal Chromosome Segment Substitution Series Derived from Japonica and Indica Cross of Rice (Oryza sativa L.). Breed. Sci. 2002, 52, 319–325. [Google Scholar] [CrossRef]
- Liu, G.; Zeng, R.; Zhu, H.; Zhang, Z.; Ding, X.; Zhao, F.; Li, W.; Zhang, G. Dynamic expression of nine QTLs for tiller number detected with single segment substitution lines in rice. Theor. Appl. Genet. 2009, 118, 443–453. [Google Scholar] [CrossRef]
- Liu, G.; Zhu, H.; Liu, S.; Zeng, R.; Zhang, Z.; Li, W.; Ding, X.; Zhao, F.; Zhang, G. Unconditional and conditional QTL mapping for the developmental behavior of tiller number in rice (Oryza sativa L.). Genetica 2010, 138, 885–893. [Google Scholar] [CrossRef]
- Liu, G.F.; Li, M.; Wen, J.; Du, Y.; Zhang, Y.M. Functional mapping of quantitative trait loci associated with rice tillering. Mol. Genet. Genom. 2010, 284, 263–271. [Google Scholar] [CrossRef]
- Yan, J.Q.; Zhu, J.; He, C.X.; Benmoussa, M.; Wu, P. Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.). Theor. Appl. Genet. 1998, 97, 267–274. [Google Scholar] [CrossRef]
- Cui, Y.; Hu, X.; Liang, G.; Feng, A.; Wang, F.; Ruan, S.; Dong, G.; Shen, L.; Zhang, B.; Chen, D.; et al. Production of novel beneficial alleles of a rice yield-related QTL by CRISPR/Cas9. Plant Biotechnol. J. 2020, 18, 1987–1989. [Google Scholar] [CrossRef]
- Ren, M.; Huang, M.; Qiu, H.; Chun, Y.; Li, L.; Kumar, A.; Fang, J.; Zhao, J.; He, H.; Li, X. Genome-Wide Association Study of the Genetic Basis of Effective Tiller Number in Rice. Rice 2021, 14, 56. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, D.; Yan, S.; Liu, S.; Liu, B.; Kang, H.; Wang, G.-L. Dissection of the Genetic Architecture of Rice Tillering using a Genome-wide Association Study. Rice 2019, 12, 43. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef]
- Shao, G.; Lu, Z.; Xiong, J.; Wang, B.; Jing, Y.; Meng, X.; Liu, G.; Ma, H.; Liang, Y.; Chen, F.; et al. Tiller Bud Formation Regulators MOC1 and MOC3 Cooperatively Promote Tiller Bud Outgrowth by Activating FON1 Expression in Rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Sun, L.; Chen, Y.; Xue, P.; Yang, Q.; Wang, B.; Yu, N.; Cao, Y.; Zhang, Y.; Gong, K.; et al. Rice dwarf and low tillering 10 (OsDLT10) regulates tiller number by monitoring auxin homeostasis. Plant Sci. 2020, 297, 110502. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.H.; Shang, F.; Lin, Q.T.; Lou, C.; Zhang, J. Tillering and panicle branching genes in rice. Gene 2014, 537, 1–5. [Google Scholar] [CrossRef]
- Yan, Y.; Ding, C.; Zhang, G.; Hu, J.; Zhu, L.; Zeng, D.; Qian, Q.; Ren, D. Genetic and environmental control of rice tillering. Crop J. 2023; in press. [Google Scholar] [CrossRef]
- Aulakh, M.S.; Wassmann, R.; Rennenberg, H. Methane transport capacity of twenty-two rice cultivars from five major Asian rice-growing countries. Agric. Ecosyst. Environ. 2002, 91, 59–71. [Google Scholar] [CrossRef]
- Chen, Y.; Lia, S.; Zhang, Y.; Li, T.; Ge, H.; Xia, S.; Gu, J.; Zhang, H.; Lü, B.; Wu, X.; et al. Rice root morphological and physiological traits interaction with rhizosphere soil and its effect on methane emissions in paddy fields. Soil Biol. Biochem. 2019, 129, 191–200. [Google Scholar] [CrossRef]
- Aulakh, M.S.; Bodenbender, J.; Wassmann, R.; Rennenberg, H. Methane transport capacity of rice plants. II. Variations among different rice cultivars and relationship with morphological characteristics. Nutr. Cycl. Agroecosyst. 2000, 58, 367–375. [Google Scholar] [CrossRef]
- Mariko, S.; Harazono, Y.; Owa, N.; Nouchi, I. Methane in flooded soil water and the emission through rice plants to the atmosphere. Environ. Exp. Bot. 1991, 31, 343–350. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, Y.; Wei, Z.; Zhu, X.; Shen, W.; Ma, J.; Lv, S.; Xu, H. The low greenhouse gas emission intensity in water-saving and drought-resistance rice in a rainfed paddy field in Southwest China. Field Crops Res. 2023, 302, 109045. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Won, Y.J.; Jeong, Y.-M.; Lee, Y.-Y.; Kang, D.-Y.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; et al. QTL mapping for pre-harvest sprouting resistance in japonica rice varieties utilizing genome re-sequencing. Mol. Genet. Genom. 2020, 295, 1129–1140. [Google Scholar] [CrossRef]
- Ji, H.; Shin, Y.; Lee, C.; Oh, H.; Yoon, I.S.; Baek, J.; Cha, Y.S.; Lee, G.S.; Kim, S.L.; Kim, K.H. Genomic Variation in Korean japonica Rice Varieties. Genes 2021, 12, 1749. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Jeong, Y.-M.; Oh, H.; Oh, J.; Kang, D.-Y.; Kim, N.; Lee, E.; Baek, J.; Kim, S.L.; Choi, I.; et al. Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties. Plants 2020, 9, 1531. [Google Scholar] [CrossRef]
- Lorieux, M. MapDisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 2012, 30, 1231–1235. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Basten, C.J.; Weir, B.S.; Zeng, Z.-B. QTL Cartographer: A Reference Manual and Tutorial for QTL Mapping; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 1996. [Google Scholar]
- Fang, F.; Ye, S.; Tang, J.; Bennett, M.J.; Liang, W. DWT1/DWL2 act together with OsPIP5K1 to regulate plant uniform growth in rice. New Phytol. 2020, 225, 1234–1246. [Google Scholar] [CrossRef]
- Choi, M.S.; Woo, M.O.; Koh, E.B.; Lee, J.; Ham, T.H.; Seo, H.S.; Koh, H.J. Teosinte Branched 1 modulates tillering in rice plants. Plant Cell Rep. 2012, 31, 57–65. [Google Scholar] [CrossRef]
- Minakuchi, K.; Kameoka, H.; Yasuno, N.; Umehara, M.; Luo, L.; Kobayashi, K.; Hanada, A.; Ueno, K.; Asami, T.; Yamaguchi, S.; et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 2010, 51, 1127–1135. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Yano, K.; Ookawa, T.; Aya, K.; Ochiai, Y.; Hirasawa, T.; Ebitani, T.; Takarada, T.; Yano, M.; Yamamoto, T.; Fukuoka, S.; et al. Isolation of a Novel Lodging Resistance QTL Gene Involved in Strigolactone Signaling and Its Pyramiding with a QTL Gene Involved in Another Mechanism. Mol. Plant 2015, 8, 303–314. [Google Scholar] [CrossRef]
- Ao, H.; Peng, S.; Zou, Y.; Tang, Q.; Visperas, R.M. Reduction of unproductive tillers did not increase the grain yield of irrigated rice. Field Crops Res. 2010, 116, 108–115. [Google Scholar] [CrossRef]
- Lyu, J.; Huang, L.; Zhang, S.; Zhang, Y.; He, W.; Zeng, P.; Zeng, Y.; Huang, G.; Zhang, J.; Ning, M.; et al. Neo-functionalization of a Teosinte branched 1 homologue mediates adaptations of upland rice. Nat. Commun. 2020, 11, 725. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, H.; Ma, B.; Liu, G.; Wang, J.; Wang, J.; Gao, R.; Li, J.; Liu, J.; Xu, J.; et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 2017, 8, 14789. [Google Scholar] [CrossRef]



| Measurement Date (DAS) | QTL Name | Chr. | Location (cm) | QTL Interval (cm) | Interval-Flanking Markers | LOD | Additive Effect | R2 | |
|---|---|---|---|---|---|---|---|---|---|
| Left | Right | ||||||||
| 14 March 2019 (41) | qTN1 | 1 | 208.1 | 204.9–208.3 | KJ01_100 | KJ01_113 | 4.9 | 0.42 | 0.036 |
| qTN3 | 3 | 132.4 | 131.3–132.6 | OU3FC_05 | KJ03_068 | 38.6 | 1.56 | 0.475 | |
| qTN7 | 7 | 117.2 | 111.9–125.3 | KJ07_071 | KJ07_084 | 8.5 | −0.65 | 0.083 | |
| 20 March 2019 (47) | qTN1.2 | 1 | 57.7 | 55.1–63.1 | KJ01_029 | KJ01_044 | 5.7 | −0.54 | 0.046 |
| qTN3 | 3 | 132.4 | 131.3–132.9 | OU3FC_05 | OU3FC_07 | 32.8 | 1.53 | 0.402 | |
| qTN5.2 | 5 | 142.4 | 138.7–144.3 | KJ05_066 | KJ05_071 | 4.3 | −0.49 | 0.043 | |
| qTN7 | 7 | 117.2 | 114.2–124.1 | KJ07_074 | KJ07_084 | 7.7 | −0.70 | 0.082 | |
| 10 June 2022 (40) | qTN1 | 1 | 208.1 | 204.5–208.4 | KJ01_112 | KJ01_113 | 4.7 | 0.31 | 0.045 |
| qTN3 | 3 | 132.4 | 131.2–132.6 | OU3FC_06 | KJ03_068 | 25.3 | 0.88 | 0.335 | |
| qTN5 | 5 | 129.8 | 125.1–132.1 | KJ05_059 | KJ05_066 | 5.5 | −0.37 | 0.060 | |
| qTN7 | 7 | 112.4 | 109.4–120.2 | KJ07_071 | KJ07_084 | 5.0 | −0.36 | 0.054 | |
| 17 June 2022 (47) | qTN3 | 3 | 132.4 | 131.1–132.6 | OU3FC_06 | KJ03_068 | 23.9 | 0.91 | 0.296 |
| qTN5 | 5 | 130.8 | 125.7–131.7 | KJ05_059 | KJ05_066 | 5.6 | −0.40 | 0.060 | |
| qTN7 | 7 | 111.4 | 106.2–121.5 | KJ07_069 | KJ07_084 | 4.7 | −0.37 | 0.046 | |
| 24 June 2022 (54) | qTN3 | 3 | 132.4 | 131.1–132.5 | OU3FC_06 | KJ03_068 | 24.7 | 0.93 | 0.302 |
| qTN5 | 5 | 130.8 | 125.3–131.7 | KJ05_059 | KJ05_066 | 5.1 | −0.39 | 0.054 | |
| qTN7 | 7 | 111.4 | 105.8–121.8 | KJ07_069 | KJ07_084 | 4.4 | −0.36 | 0.042 | |
| qTN11 | 11 | 3.0 | 0–14.0 | KJ11_004 | KJ11_008 | 3.4 | 0.33 | 0.040 | |
| 1 July 2022 (61) | qTN3 | 3 | 132.4 | 131.2–132.6 | OU3FC_06 | KJ03_068 | 21.6 | 1.12 | 0.188 |
| qTN5 | 5 | 131.0 | 128.3–131.9 | KJ05_059 | KJ05_066 | 6.0 | −0.70 | 0.078 | |
| Trait | QTL Name | Chr. | Location (cm) | QTL Interval (cm) | Interval-Flanking Markers | LOD | Additive Effect | R2 | |
|---|---|---|---|---|---|---|---|---|---|
| Left | Right | ||||||||
| CL | qCL1 | 1 | 209.4 | 208.4–211.5 | KJ01_113 | KJ01_115 | 27.0 | −5.26 | 0.437 |
| qCL2 | 2 | 99.4 | 92.7–100.4 | KJ02_064 | KJ02_068 | 4.4 | −1.73 | 0.048 | |
| qCL4 | 4 | 66.1 | 55.3–76.1 | KJ04_059 | OUC04_1635 | 3.3 | 1.94 | 0.058 | |
| PL | qPL1 | 1 | 207.9 | 204.9–208.1 | KJ01_099 | KJ01_112 | 6.9 | −0.65 | 0.110 |
| qPL3 | 3 | 2.0 | 0–3.9 | KJ03_004 | KJ03_008 | 4.0 | −0.48 | 0.059 | |
| qPL6 | 6 | 54.7 | 54.0–56.7 | KJ06_028 | KJ06_039 | 5.6 | 0.60 | 0.097 | |
| qPL10 | 10 | 0.0 | 0–2.0 | KJ10_006 | OUC10_2109 | 4.0 | 0.49 | 0.059 | |
| PN | qPN3 | 3 | 128.7 | 125.5–131.4 | OU3FC_01 | KJ03_068 | 4.9 | 0.39 | 0.096 |
| qPN5 | 5 | 122.8 | 117.5–128.2 | KJ05_058 | KJ05_063 | 3.3 | −0.37 | 0.092 | |
| qPN8 | 8 | 54.6 | 53.4–55.6 | KJ08_040 | KJ08_044 | 3.4 | 1.86 | 0.119 | |
| CD | qCD3 | 3 | 130.7 | 126.3–132.7 | OU3FC_03 | OU3FC_07 | 4.2 | −0.12 | 0.071 |
| qCD8 | 8 | 67.7 | 65.4–75.5 | KJ08_075 | OUC08_13457 | 6.3 | −0.17 | 0.144 | |
| GNP | qGNP1 | 1 | 207.9 | 202.5–208.1 | KJ01_100 | KJ01_112 | 9.9 | −5.23 | 0.146 |
| qGNP3 | 3 | 162.3 | 158.1–163.3 | OUC03_1163 | OUC03_1175 | 7.6 | 4.73 | 0.118 | |
| qGNP5 | 5 | 131.7 | 129.4–134.1 | KJ05_065 | KJ05_066 | 4.5 | 3.41 | 0.060 | |
| qGNP12 | 12 | 93.8 | 89.7–97.4 | KJ12_060 | KJ12_061 | 8.5 | −4.99 | 0.134 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, D.-K.; Choi, I.; Won, Y.J.; Shin, Y.; Cheon, K.-S.; Oh, H.; Lee, C.; Lee, S.; Cho, M.H.; Jun, S.; et al. QTL Mapping of Tiller Number in Korean Japonica Rice Varieties. Genes 2023, 14, 1593. https://doi.org/10.3390/genes14081593
Yoon D-K, Choi I, Won YJ, Shin Y, Cheon K-S, Oh H, Lee C, Lee S, Cho MH, Jun S, et al. QTL Mapping of Tiller Number in Korean Japonica Rice Varieties. Genes. 2023; 14(8):1593. https://doi.org/10.3390/genes14081593
Chicago/Turabian StyleYoon, Dong-Kyung, Inchan Choi, Yong Jae Won, Yunji Shin, Kyeong-Seong Cheon, Hyoja Oh, Chaewon Lee, Seoyeon Lee, Mi Hyun Cho, Soojin Jun, and et al. 2023. "QTL Mapping of Tiller Number in Korean Japonica Rice Varieties" Genes 14, no. 8: 1593. https://doi.org/10.3390/genes14081593
APA StyleYoon, D.-K., Choi, I., Won, Y. J., Shin, Y., Cheon, K.-S., Oh, H., Lee, C., Lee, S., Cho, M. H., Jun, S., Kim, Y., Kim, S. L., Baek, J., Jeong, H., Lyu, J. I., Lee, G.-S., Kim, K.-H., & Ji, H. (2023). QTL Mapping of Tiller Number in Korean Japonica Rice Varieties. Genes, 14(8), 1593. https://doi.org/10.3390/genes14081593







