Multiple Sclerosis Heritability Estimation on Sardinian Ascertained Extended Families Using Bayesian Liability Threshold Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Sardinian Families Ascertainment
2.2. Statistical Analysis
2.2.1. Model Specification
2.2.2. Implementing Bayesian-LTMH
3. Results and Discussion
3.1. Sample Description
3.2. Bayesian-LTMH Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Visscher, P.M.; Hill, W.G.; Wray, N.R. Heritability in the genomics era—Concepts and misconceptions. Nat. Rev. Genet. 2008, 9, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Kempthorne, O. The correlation between relatives on the supposition of mendelian inheritance. Am. J. Hum. Genet. 1968, 20, 402. [Google Scholar]
- Egeland, J. Heritability and Etiology: Heritability estimates can provide causally relevant information. Pers. Individ. Dif. 2023, 200, 111896. [Google Scholar] [CrossRef]
- Athanasiadis, G.; Speed, D.; Andersen, M.K.; Appel, E.V.R.; Grarup, N.; Brandslund, I.; Jørgensen, M.E.; Larsen, C.V.L.; Bjerregaard, P.; Hansen, T.; et al. Estimating narrow-sense heritability using family data from admixed populations. Heredity 2020, 124, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, R.; Spicer, R.; Muthukrishna, M. Cultural evolution of genetic heritability. Behav. Brain Sci. 2022, 45, e152. [Google Scholar] [CrossRef]
- Pearson, C.H. Is heritability explanatorily useful? Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2007, 38, 270–288. [Google Scholar] [CrossRef]
- Bourrat, P. Heritability, causal influence and locality. Synthese 2021, 198, 6689–6715. [Google Scholar] [CrossRef]
- Tal, O. From heritability to probability. Biol. Philos. 2009, 24, 81–105. [Google Scholar] [CrossRef]
- Milo, R.; Kahana, E. Multiple sclerosis: Geoepidemiology, genetics and the environment. Autoimmun. Rev. 2010, 9, A387–A394. [Google Scholar] [CrossRef]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Schriml, L.M.; Lichenstein, R.; Bisordi, K.; Bearer, C.; Baron, J.A.; Greene, C. Modeling the enigma of complex disease etiology. J. Transl. Med. 2023, 21, 148. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, N.; Razavi, S.; Nikzad, E. Multiple Sclerosis: Pathogenesis, Symptoms, Diagnoses andCell-Based Therapy. Cell J. 2017, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Fazia, T.; Marzanati, D.; Carotenuto, A.L.; Beecham, A.; Hadjixenofontos, A.; McCauley, J.L.; Saddi, V.; Piras, M.; Bernardinelli, L.; Gentilini, D. Homozygosity Haplotype and Whole-Exome Sequencing Analysis to Identify Potentially Functional Rare Variants Involved in Multiple Sclerosis among Sardinian Families. Curr. Issues Mol. Biol. 2021, 43, 1778–1793. [Google Scholar] [CrossRef] [PubMed]
- Fazia, T.; Baldrighi, G.N.; Nova, A.; Bernardinelli, L. A systematic review of Mendelian randomization studies on multiple sclerosis. Eur. J. Neurosci. 2023. [Google Scholar] [CrossRef] [PubMed]
- Waubant, E.; Lucas, R.; Mowry, E.; Graves, J.; Olsson, T.; Alfredsson, L.; Langer-Gould, A. Environmental and genetic risk factors for MS: An integrated review. Ann. Clin. Transl. Neurol. 2019, 6, 1905–1922. [Google Scholar] [CrossRef]
- Amato, M.P.; Derfuss, T.; Hemmer, B.; Liblau, R.; Montalban, X.; Soelberg Sørensen, P.; Miller, D.H.; Alfredsson, L.; Aloisi, F.; Ascherio, A.; et al. Environmental modifiable risk factors for multiple sclerosis: Report from the 2016 ECTRIMS focused workshop. Mult. Scler. 2018, 24, 590–603. [Google Scholar] [CrossRef]
- Patsopoulos, N.A.; Baranzini, S.E.; Santaniello, A.; Shoostari, P.; Cotsapas, C.; Wong, G.; Beecham, A.H.; James, T.; Replogle, J.; Vlachos, I.S.; et al. Multiple Sclerosis Genomic Map implicates peripheral immune cells & microglia in susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef]
- Mitrovič, M.; Patsopoulos, N.A.; Beecham, A.H.; Dankowski, T.; Goris, A.; Dubois, B.; D’hooghe, M.B.; Lemmens, R.; Van Damme, P.; Søndergaard, H.B.; et al. Low-Frequency and Rare-Coding Variation Contributes to Multiple Sclerosis Risk. Cell 2020, 180, 403. [Google Scholar] [CrossRef]
- Fazia, T.; Pastorino, R.; Foco, L.; Han, L.; Abney, M.; Beecham, A.; Hadjixenofontos, A.; Guo, H.; Gentilini, D.; Papachristou, C.; et al. Investigating multiple sclerosis genetic susceptibility on the founder population of east-central Sardinia via association and linkage analysis of immune-related loci. Mult. Scler. 2018, 24, 1815–1824. [Google Scholar] [CrossRef]
- Fagnani, C.; Neale, M.C.; Nisticò, L.; Stazi, M.A.; Ricigliano, V.A.; Buscarinu, M.C.; Salvetti, M.; Ristori, G. Twin studies in multiple sclerosis: A meta-estimation of heritability and environmentality. Mult. Scler. 2015, 21, 1404–1413. [Google Scholar] [CrossRef]
- Ristori, G.; Cannoni, S.; Stazi, M.A.; Vanacore, N.; Cotichini, R.; Alfò, M.; Pugliatti, M.; Sotgiu, S.; Solaro, C.; Bomprezzi, R.; et al. Multiple Sclerosis in Twins from Continental Italy and Sardinia: A Nationwide Study. Ann. Neurol. 2005, 59, 27–34. [Google Scholar] [CrossRef]
- Kruuk, L.E.B.; Hadfield, J.D. How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 2007, 20, 1890–1903. [Google Scholar] [CrossRef] [PubMed]
- Dick, D.M. Shared Environment. Encycl. Stat. Behav. Sci. 2005, 4, 1828–1830. [Google Scholar] [CrossRef]
- Kendler, K.S.; Ohlsson, H.; Lichtenstein, P.; Sundquist, J.; Sundquist, K. The Nature of the Shared Environment. Behav. Genet. 2019, 49, 1–10. [Google Scholar] [CrossRef]
- Pittner, K.; Bakermans-Kranenburg, M.J.; Alink, L.R.A.; Buisman, R.S.M.; van den Berg, L.J.M.; van den Block, L.H.C.D.; Voorthuis, A.; Elzinga, B.M.; Lindenberg, J.; Tollenaar, M.S.; et al. Estimating the Heritability of Experiencing Child Maltreatment in an Extended Family Design. Child Maltreat. 2020, 25, 289. [Google Scholar] [CrossRef]
- De Villemereuil, P.; Gimenez, O.; Doligez, B. Comparing parent–offspring regression with frequentist and Bayesian animal models to estimate heritability in wild populations: A simulation study for Gaussian and binary traits. Methods Ecol. Evol. 2013, 4, 260–275. [Google Scholar] [CrossRef]
- Park, S.; Lee, S.; Lee, Y.; Herold, C.; Hooli, B.; Mullin, K.; Park, T.; Park, C.; Bertram, L.; Lange, C.; et al. Adjusting heterogeneous ascertainment bias for genetic association analysis with extended families. BMC Med. Genet. 2015, 16, 62. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hawkes, C.H.; Macgregor, A.J. Twin studies and the heritability of MS: A conclusion. Mult. Scler. 2009, 15, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kwak, S.H.; Won, S. Heritability estimation of dichotomous phenotypes using a liability threshold model on ascertained family-based samples. Genet. Epidemiol. 2019, 43, 761–775. [Google Scholar] [CrossRef]
- Louis, T.A. Finding the Observed Information Matrix When Using the EM Algorithm. J. R. Stat. Soc. Ser. B 1982, 44, 226–233. [Google Scholar] [CrossRef]
- Xu, C.; Baines, P.D.; Wang, J.L. Standard error estimation using the EM algorithm for the joint modeling of survival and longitudinal data. Biostatistics 2014, 15, 731. [Google Scholar] [CrossRef] [PubMed]
- Sofer, T. Confidence intervals for heritability via Haseman-Elston regression. Stat. Appl. Genet. Mol. Biol. 2017, 16, 259. [Google Scholar] [CrossRef] [PubMed]
- Granieri, E.; Casetta, I.; Govoni, V.; Tola, M.R.; Marchi, D.; Murgia, S.B.; Ticca, A.; Pugliatti, M.; Murgia, B.; Rosati, G. The increasing incidence and prevalence of MS in a Sardinian province. Neurology 2000, 55, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Sotgiu, S.; Pugliatti, M.; Sotgiu, A.; Sanna, A.; Rosati, G. Review: Does the “Hygiene Hypothesis” Provide an Explanation for the High Prevalence of Multiple Sclerosis in Sardinia? Autoimmunity 2009, 36, 257–260. [Google Scholar] [CrossRef]
- Sotgiu, S.; Pugliatti, M.; Sanna, A.; Sotgiu, A.; Castigli, P.; Solinas, G.; Dolei, A.; Serra, C.; Bonetti, B.; Rosati, G. Multiple sclerosis complexity in selected populations: The challenge of Sardinia, insular Italy. Eur. J. Neurol. 2002, 9, 329–341. [Google Scholar] [CrossRef]
- Matveeva, O.; Bogie, J.F.J.; Hendriks, J.J.A.; Linker, R.A.; Haghikia, A.; Kleinewietfeld, M. Western lifestyle and immunopathology of multiple sclerosis. Ann. N. Y. Acad. Sci. 2018, 1417, 71. [Google Scholar] [CrossRef]
- Tognotti, E. Program to Eradicate Malaria in Sardinia, 1946–1950. Emerg. Infect. Dis. 2009, 15, 1460. [Google Scholar] [CrossRef]
- Riedl, B.; Beckmann, T.; Neundõrfer, B.; Handwerker, H.O.; Birklein, F. Multiple sclerosis epidemiology in Sardinia: Evidence for a true increasing risk. Acta Neurol. Scand. 2001, 103, 20–26. [Google Scholar] [CrossRef]
- Casetta, I.; Granieri, E.; Marchi, D.; Murgia, S.B.; Tola, M.R.; Ticca, A.; Lauria, G.; Govoni, V.; Murgia, B.; Pugliatti, M. An epidemiological study of multiple sclerosis in central Sardinia, Italy. Acta Neurol. Scand. 1998, 98, 391–394. [Google Scholar] [CrossRef]
- Poser, C.M.; Paty, D.W.; Scheinberg, L.; McDonald, W.I.; Davis, F.A.; Ebers, G.C.; Johnson, K.P.; Sibley, W.A.; Silberberg, D.H.; Tourtellotte, W.W. New diagnostic criteria for multiple sclerosis: Guidelines for research protocols. Ann. Neurol. 1983, 13, 227–231. [Google Scholar] [CrossRef]
- Hujoel, M.L.A.; Gazal, S.; Loh, P.R.; Patterson, N.; Price, A.L. Liability threshold modeling of case–control status and family history of disease increases association power. Nat. Genet. 2020, 52, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Montomoli, C.; Allemani, C.; Solinas, G.; Motta, G.; Bernardinelli, L.; Clemente, S.; Murgia, B.S.; Ticca, A.F.; Musu, L.; Piras, M.L.; et al. An ecologic study of geographical variation in multiple sclerosis risk in central Sardinia, Italy. Neuroepidemiology 2002, 21, 187–193. [Google Scholar] [CrossRef]
- Urru, S.A.M.; Antonelli, A.; Sechi, G.M. Prevalence of multiple sclerosis in Sardinia: A systematic cross-sectional multi-source survey. Mult. Scler. J. 2020, 26, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R. Concepts and Misconceptions about the Polygenic Additive Model Applied to Disease. Hum. Hered. 2015, 80, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Ebers, G.C.; Sadovnick, A.D.; Dyment, D.A.; Yee, I.M.L.; Willer, C.J.; Risch, N. Parent-of-origin effect in multiple sclerosis: Observations in half-siblings. Lancet 2004, 363, 1773–1774. [Google Scholar] [CrossRef]
- Hoppenbrouwers, I.A.; Liu, F.; Aulchenko, Y.S.; Ebers, G.C.; Oostra, B.A.; Van Duijn, C.M.; Hintzen, R.Q. Maternal transmission of multiple sclerosis in a dutch population. Arch. Neurol. 2008, 65, 345–348. [Google Scholar] [CrossRef]
- Benchek, P.H.; Morris, N.J. How meaningful are heritability estimates of liability? Hum. Genet. 2013, 132, 1351–1360. [Google Scholar] [CrossRef]
- Gjessing, H.K.; Lie, R.T. Biometrical modelling in genetics: Are complex traits too complex? Stat. Methods Med. Res. 2008, 17, 75–96. [Google Scholar] [CrossRef]
- de Villemereuil, P.; Morrissey, M.B.; Nakagawa, S.; Schielzeth, H. Fixed-effect variance and the estimation of repeatabilities and heritabilities: Issues and solutions. J. Evol. Biol. 2018, 31, 621–632. [Google Scholar] [CrossRef]
- Nakagawa, S.; Schielzeth, H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013, 4, 133–142. [Google Scholar] [CrossRef]
- Almasy, L.; Towne, B.; Peterson, C.; Blangero, J. Detecting genotype x age interaction. Genet. Epidemiol. 2001, 21 (Suppl. 1), S819–S824. [Google Scholar] [CrossRef]
- Poveda, A.; Chen, Y.; Brändström, A.; Engberg, E.; Hallmans, G.; Johansson, I.; Renström, F.; Kurbasic, A.; Franks, P.W. The heritable basis of gene-environment interactions in cardiometabolic traits. Diabetologia 2017, 60, 442–452. [Google Scholar] [CrossRef]
- Paap, R. What are the advantages of MCMC based inference in latent variable models? Stat. Neerl. 2002, 56, 2–22. [Google Scholar] [CrossRef]
- Dunson, D.B. Commentary: Practical Advantages of Bayesian Analysis of Epidemiologic Data. Am. J. Epidemiol. 2001, 153, 1222–1226. [Google Scholar] [CrossRef]
- Luengo, D.; Martino, L.; Bugallo, M.; Elvira, V.; Särkkä, S. A survey of Monte Carlo methods for parameter estimation. EURASIP J. Adv. Signal Process. 2020, 2020, 25. [Google Scholar] [CrossRef]
- Tang, Y. Beyond EM: A faster Bayesian linear regression algorithm without matrix inversions. Neurocomputing 2020, 378, 435–440. [Google Scholar] [CrossRef]
- Hamra, G.; MacLehose, R.; Richardson, D. Markov Chain Monte Carlo: An introduction for epidemiologists. Int. J. Epidemiol. 2013, 42, 627. [Google Scholar] [CrossRef] [PubMed]
- Elston, R.C.; Olson, J.M.; Palmer, L. Biostatistical Genetics and Genetic Epidemiology; John Wiley & Sons: Hoboken, NJ, USA, 2002; p. 831. [Google Scholar]
- Carpenter, B.; Gelman, A.; Hoffman, M.D.; Lee, D.; Goodrich, B.; Betancourt, M.; Brubaker, M.A.; Guo, J.; Li, P.; Riddell, A. Stan: A Probabilistic Programming Language. J. Stat. Softw. 2017, 76, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Betancourt, M. A Conceptual Introduction to Hamiltonian Monte Carlo. arXiv 2018, arXiv:1701.02434. [Google Scholar]
- Jonah Gabry and Rok Cesnovar cmdstanr: R Interface to “CmdStan”. 2021.
- Sotgiu, S.; Angius, A.; Embry, A.; Rosati, G.; Musumeci, S. Hygiene hypothesis: Innate immunity, malaria and multiple sclerosis. Med. Hypotheses 2008, 70, 819–825. [Google Scholar] [CrossRef]
- Handel, A.E.; Williamson, A.J.; Disanto, G.; Handunnetthi, L.; Giovannoni, G.; Ramagopalan, S.V. An updated meta-analysis of risk of multiple sclerosis following infectious mononucleosis. PLoS ONE 2010, 5, e12496. [Google Scholar] [CrossRef] [PubMed]
- Mameli, G.; Cossu, D.; Cocco, E.; Masala, S.; Frau, J.; Marrosu, M.G.; Sechi, L.A. EBNA-1 IgG titers in Sardinian multiple sclerosis patients and controls. J. Neuroimmunol. 2013, 264, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Alfredsson, L.; Olsson, T. Lifestyle and Environmental Factors in Multiple Sclerosis. Cold Spring Harb. Perspect. Med. 2019, 9, a028944. [Google Scholar] [CrossRef] [PubMed]
- Ascherio, A.; Munger, K.L.; Lünemann, J.D. The initiation and prevention of multiple sclerosis. Nat. Rev. Neurol. 2012, 8, 602–612. [Google Scholar] [CrossRef]
- Puthenparampil, M.; Perini, P.; Bergamaschi, R.; Capobianco, M.; Filippi, M.; Gallo, P. Multiple sclerosis epidemiological trends in Italy highlight the environmental risk factors. J. Neurol. 2022, 269, 1817–1824. [Google Scholar] [CrossRef]
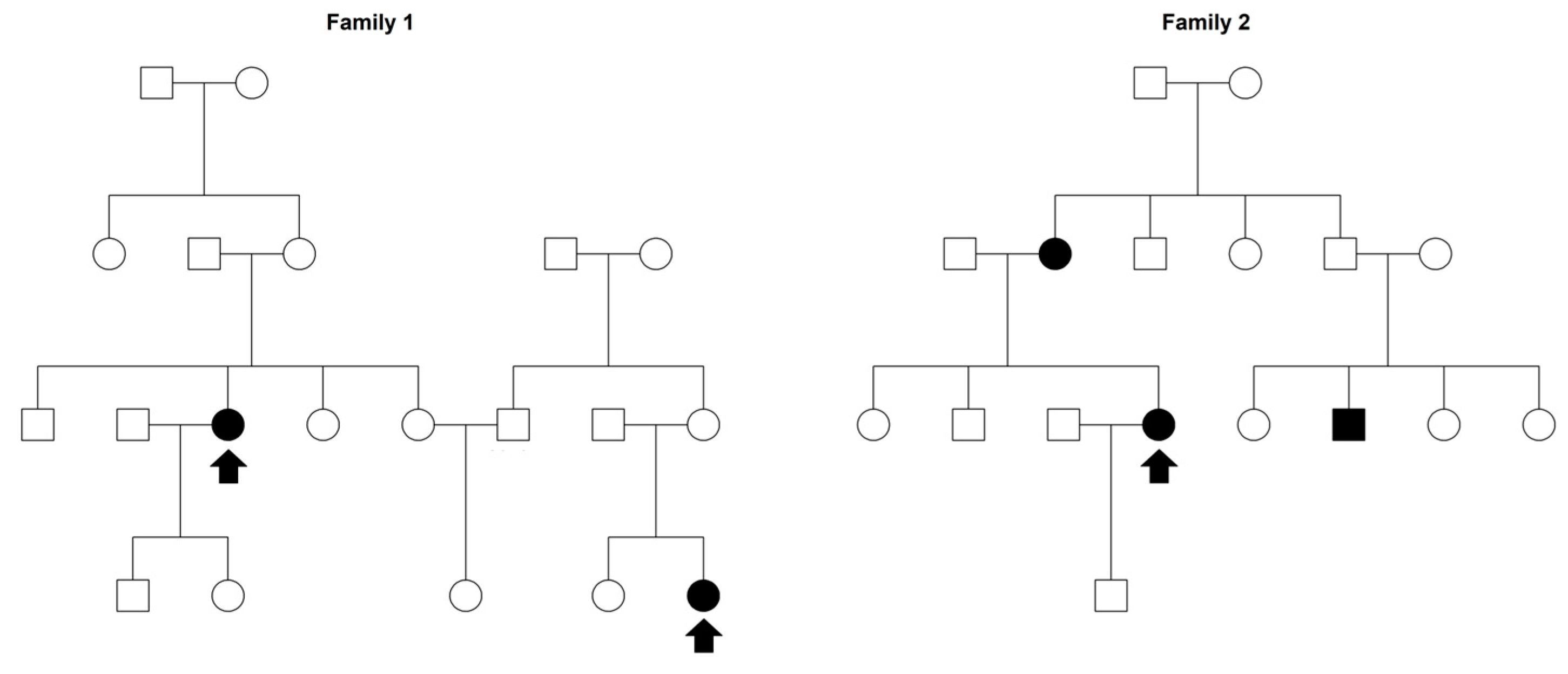
| Family | Individuals N (%) 1 | Probands N | Females N (%) 2 | MS Cases N (%) 2 |
|---|---|---|---|---|
| 1 | 65 (8%) | 6 | 37 (57%) | 6 (9%) |
| 2 | 35 (4%) | 4 | 20 (57%) | 5 (14%) |
| 3 | 70 (9%) | 7 | 45 (64%) | 9 (13%) |
| 4 | 66 (8%) | 8 | 37 (56%) | 10 (15%) |
| 5 | 12 (2%) | 2 | 6 (50%) | 3 (25%) |
| 6 | 16 (2%) | 2 | 7 (44%) | 2 (13%) |
| 7 | 43 (5%) | 5 | 24 (56%) | 5 (12%) |
| 8 | 33 (4%) | 5 | 16 (48%) | 6 (18%) |
| 9 | 17 (2%) | 2 | 10 (59%) | 2 (12%) |
| 10 | 20 (3%) | 2 | 13 (65%) | 3 (15%) |
| 11 | 15 (2%) | 1 | 8 (53%) | 3 (20%) |
| 12 | 33 (4%) | 5 | 17 (52%) | 6 (18%) |
| 13 | 17 (2%) | 2 | 11 (65%) | 3 (18%) |
| 14 | 51 (6%) | 6 | 24 (47%) | 12 (24%) |
| 15 | 25 (3%) | 3 | 16 (64%) | 3 (12%) |
| 16 | 44 (6%) | 5 | 24 (55%) | 8 (18%) |
| 17 | 19 (2%) | 2 | 12 (63%) | 2 (11%) |
| 18 | 16 (2%) | 2 | 8 (50%) | 2 (13%) |
| 19 | 22 (3%) | 3 | 13 (59%) | 3 (14%) |
| 20 | 27 (3%) | 2 | 16 (59%) | 2 (7%) |
| 21 | 28 (4%) | 1 | 13 (46%) | 2 (7%) |
| 22 | 16 (2%) | 2 | 7 (44%) | 4 (25%) |
| 23 | 7 (1%) | 1 | 3 (43%) | 1 (14%) |
| 24 | 93 (12%) | 11 | 48 (52%) | 16 (17%) |
| Total | 790 | 89 | 435 (55%) | 118 (15%) |
| MS Course ° | N (%) | Females (%) | Age MS Onset Mean (SD) | Year MS Onset Mean (SD) |
|---|---|---|---|---|
| RRMS | 58 (49%) | 41 (71%) | 28.45 (9.49) | 1990 (10.09) |
| SPMS | 27 (23%) | 14 (52%) | 28.89 (8.87) | 1983 (9.64) |
| PPMS | 1 (1%) | 1 (100%) | 45.00 | 1995 |
| Unknown | 32 (27%) | 20 (63%) | N/A | N/A |
| Total | 118 | 76 (64%) | 28.64 (9.06) * | 1988 (10.88) * |
| Kinship Relationship | N (%) * |
|---|---|
| First degree | 20 (8%) |
| Parent–offspring | 9 |
| Mother | 6 |
| Father | 3 |
| Sibling | 13 |
| Second degree | 9 (4%) |
| Uncle/aunt–nephew/niece | 8 |
| Grandparent–grandchild | 1 |
| Third degree | 16 (7%) |
| Cousins | 15 |
| Grand-grandparent–grand-grandchild | 1 |
| Fourth degree | 17 (7%) |
| Over the fourth degree | 176 (74%) |
| Total | 238 |
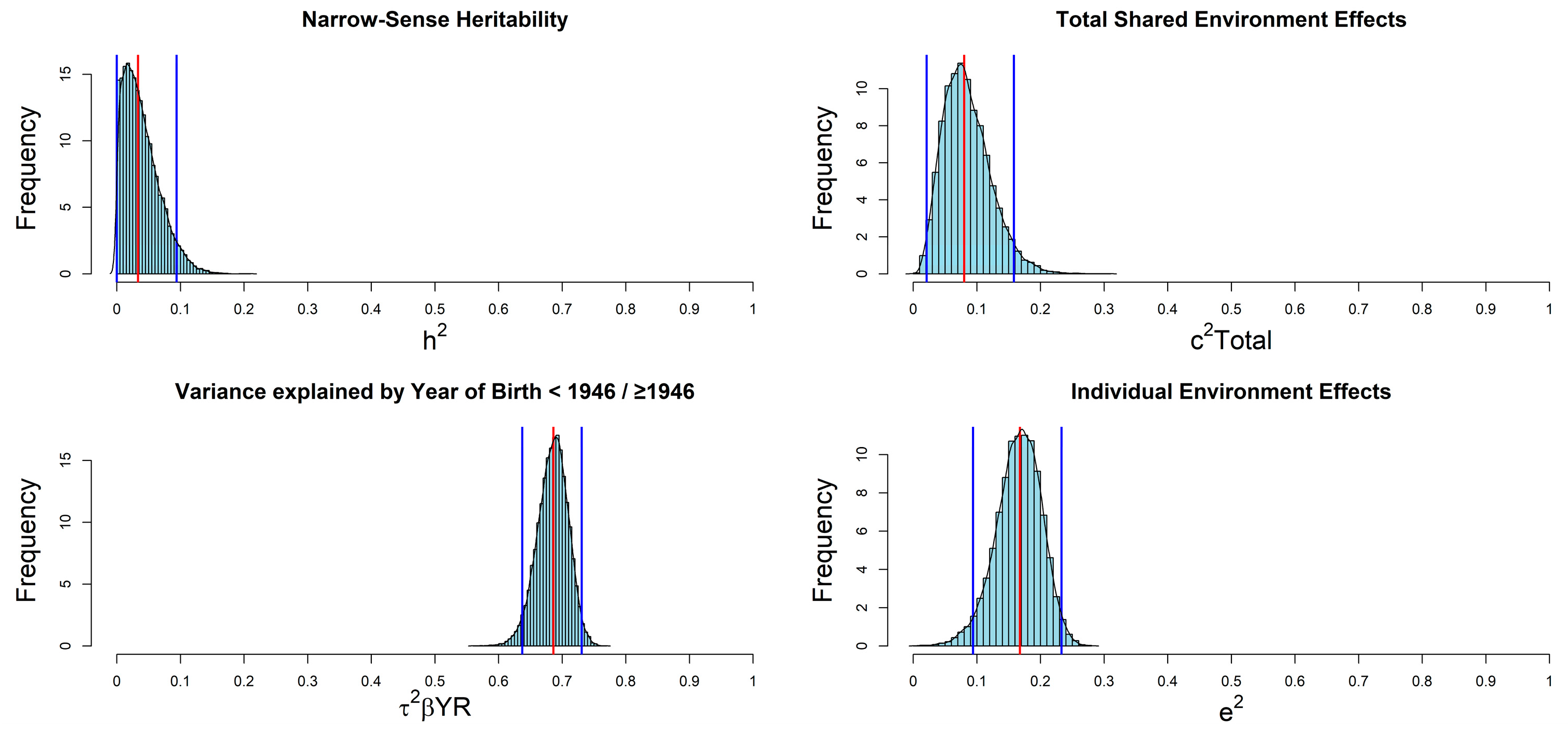
| Parameter | Median | SD 1 | HPD 95% CI 1 |
|---|---|---|---|
| h2 | 0.033 | 0.028 | 0.000, 0.094 |
| c2Sibs | 0.033 | 0.016 | 0.007, 0.067 |
| c2Mother–Sibs | 0.012 | 0.012 | 0.000, 0.039 |
| c2Father–Sibs | 0.013 | 0.013 | 0.000, 0.040 |
| c2Spouses | 0.014 | 0.017 | 0.000, 0.051 |
| c2Total | 0.080 | 0.037 | 0.021, 0.158 |
| e2 | 0.168 | 0.036 | 0.094, 0.233 |
| τ2βSEX,YR | 0.712 | 0.020 | 0.673, 0.749 |
| τ2βSEX | 0.009 | 0.008 | 0.000, 0.027 |
| τ2βYR | 0.686 | 0.024 | 0.637, 0.731 |
| 2cov°βSEX,YR | 0.015 | 0.007 | 0.003, 0.028 |
| βSEX(Females vs. Males) | 0.355 | 0.157 | 0.057, 0.679 |
| βYR(≥1946 vs. <1946) | 3.173 | 0.155 | 2.869, 3.477 |
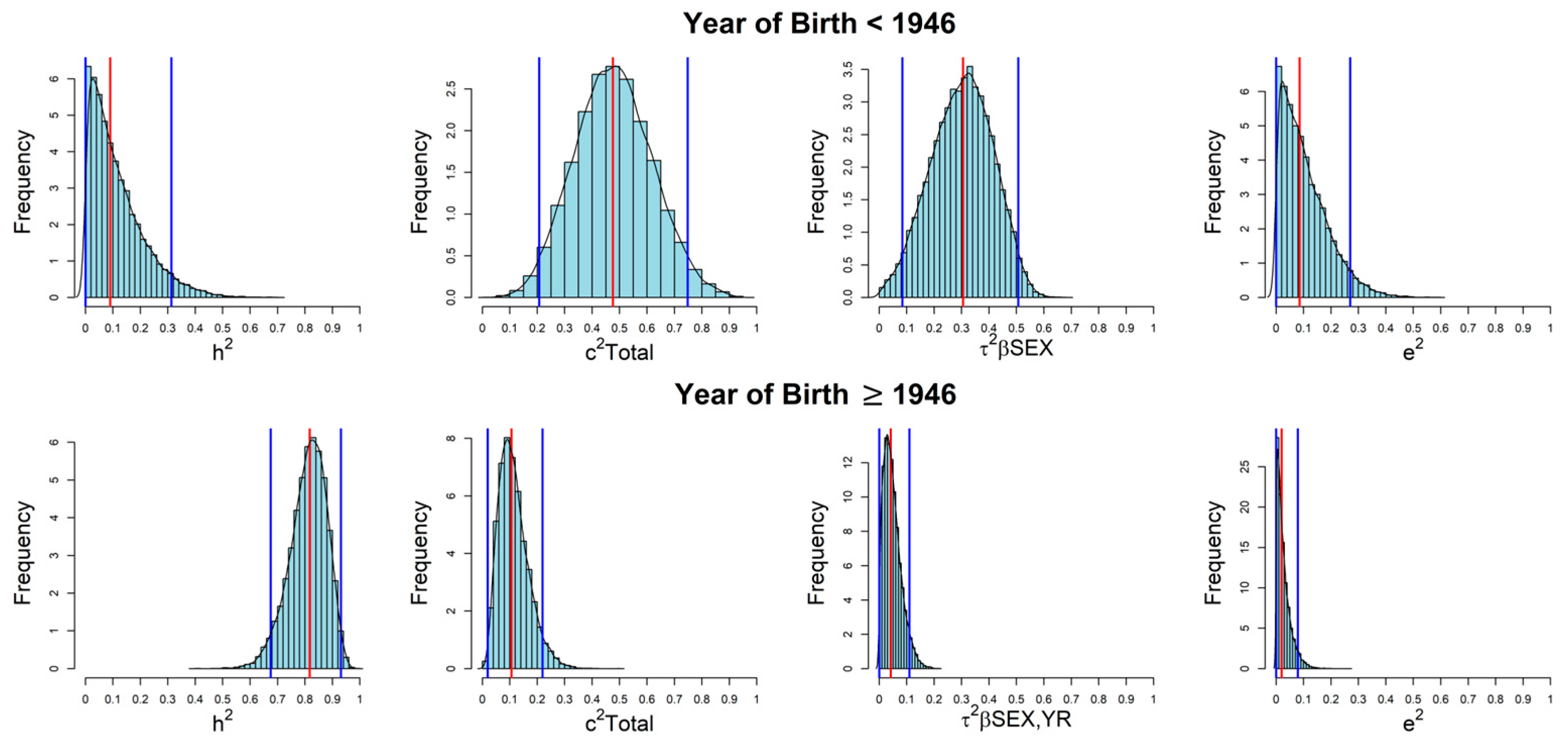
| Year of Birth < 1946 | Year of Birth ≥ 1946 | |||||
|---|---|---|---|---|---|---|
| Parameter | Median | SD 1 | 95% HPD CI 1 | Median | SD 1 | 95% HPD CI 1 |
| h2 | 0.090 | 0.100 | 0.000, 0.312 | 0.818 | 0.068 | 0.679, 0.937 |
| c2Sibs | 0.223 | 0.100 | 0.055, 0.433 | 0.045 | 0.030 | 0.004, 0.109 |
| c2Mother–Sibs | 0.061 | 0.058 | 0.000, 0.185 | 0.013 | 0.016 | 0.000, 0.050 |
| c2Father–Sibs | 0.049 | 0.051 | 0.000, 0.163 | 0.014 | 0.017 | 0.000, 0.054 |
| c2Spouses | 0.085 | 0.083 | 0.000, 0.297 | 0.019 | 0.026 | 0.000, 0.078 |
| c2Total | 0.477 | 0.142 | 0.199, 0.750 | 0.105 | 0.056 | 0.019, 0.222 |
| e2 | 0.086 | 0.083 | 0.000, 0.265 | 0.021 | 0.025 | 0.000, 0.078 |
| τ2βSEX,YR | N/A 1 | N/A 1 | N/A 1 | 0.042 | 0.032 | 0.000, 0.109 |
| τ2βSEX | 0.304 | 0.112 | 0.079, 0.506 | 0.005 | 0.013 | 0.000, 0.035 |
| τ2βYR | N/A 1 | N/A 1 | N/A 1 | 0.032 | 0.030 | 0.001, 0.095 |
| 2cov°βSEX,YR | N/A 1 | N/A 1 | N/A 1 | 0.000 | 0.001 | −0.001, 0.001 |
| βSEX(Females vs. Males) | 1.322 | 0.368 | 0.586, 2.023 | 0.104 | 0.177 | −0.246, 0.448 |
| βYR(10 years increase) | N/A 1 | N/A 1 | N/A 1 | 0.186 | 0.089 | 0.012, 0.362 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nova, A.; Fazia, T.; Saddi, V.; Piras, M.; Bernardinelli, L. Multiple Sclerosis Heritability Estimation on Sardinian Ascertained Extended Families Using Bayesian Liability Threshold Model. Genes 2023, 14, 1579. https://doi.org/10.3390/genes14081579
Nova A, Fazia T, Saddi V, Piras M, Bernardinelli L. Multiple Sclerosis Heritability Estimation on Sardinian Ascertained Extended Families Using Bayesian Liability Threshold Model. Genes. 2023; 14(8):1579. https://doi.org/10.3390/genes14081579
Chicago/Turabian StyleNova, Andrea, Teresa Fazia, Valeria Saddi, Marialuisa Piras, and Luisa Bernardinelli. 2023. "Multiple Sclerosis Heritability Estimation on Sardinian Ascertained Extended Families Using Bayesian Liability Threshold Model" Genes 14, no. 8: 1579. https://doi.org/10.3390/genes14081579
APA StyleNova, A., Fazia, T., Saddi, V., Piras, M., & Bernardinelli, L. (2023). Multiple Sclerosis Heritability Estimation on Sardinian Ascertained Extended Families Using Bayesian Liability Threshold Model. Genes, 14(8), 1579. https://doi.org/10.3390/genes14081579







