Genome-Wide Identification and Characterization of the Biosynthesis of the Polyamine Gene Family in Citrus unshiu
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Growth Condition and Transcriptome Analysis
2.2. Database Search, Sequence Retrieval, and Physiochemical Properties
2.3. Multiple Sequence Alignment, Phylogenetic Analysis, and the Biosynthesis of PA Gene Orthologs in Arabidopsis
2.4. Conserved Motifs, Cis-Element, and Gene Structural Analysis
2.5. Scaffold Location, Synteny Analysis, and Gene Duplication Event
2.6. Effect of Peel Roughing Disease Stress on C. unshiu PA Biosynthestic Genes
2.7. Citrus unshiu Plant under Seedling Etiolation
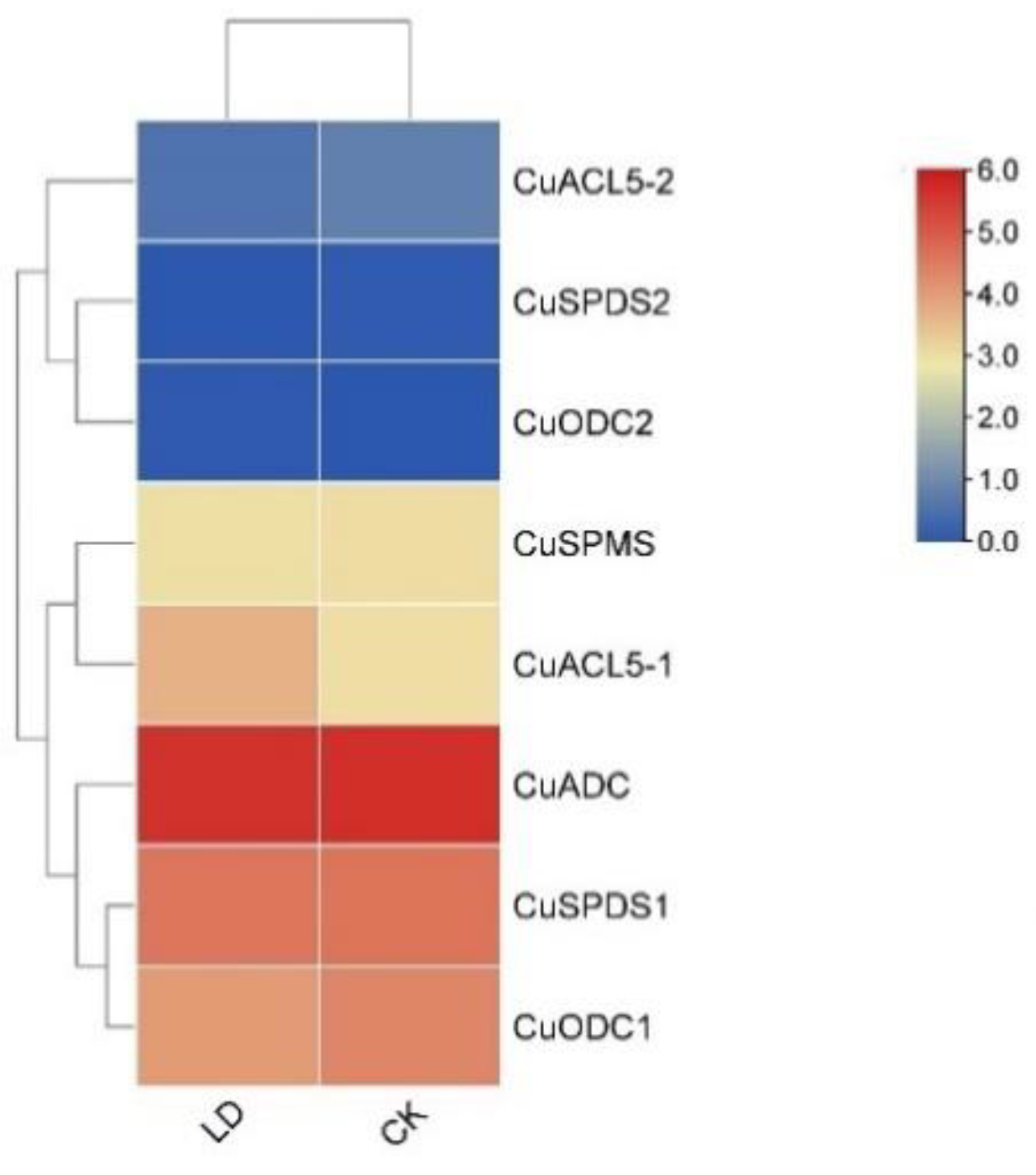
2.8. Light Drought Effect on C. unshiu PA Biosynthetic Genes during Induction of Flowering Branches
3. Results
3.1. Identification and Characterization of the Biosynthesis of the PA Gene Family in C. unshiu
3.2. Comparative Phylogenetic and Orthologs Analysis of PA Biosynthetic Genes
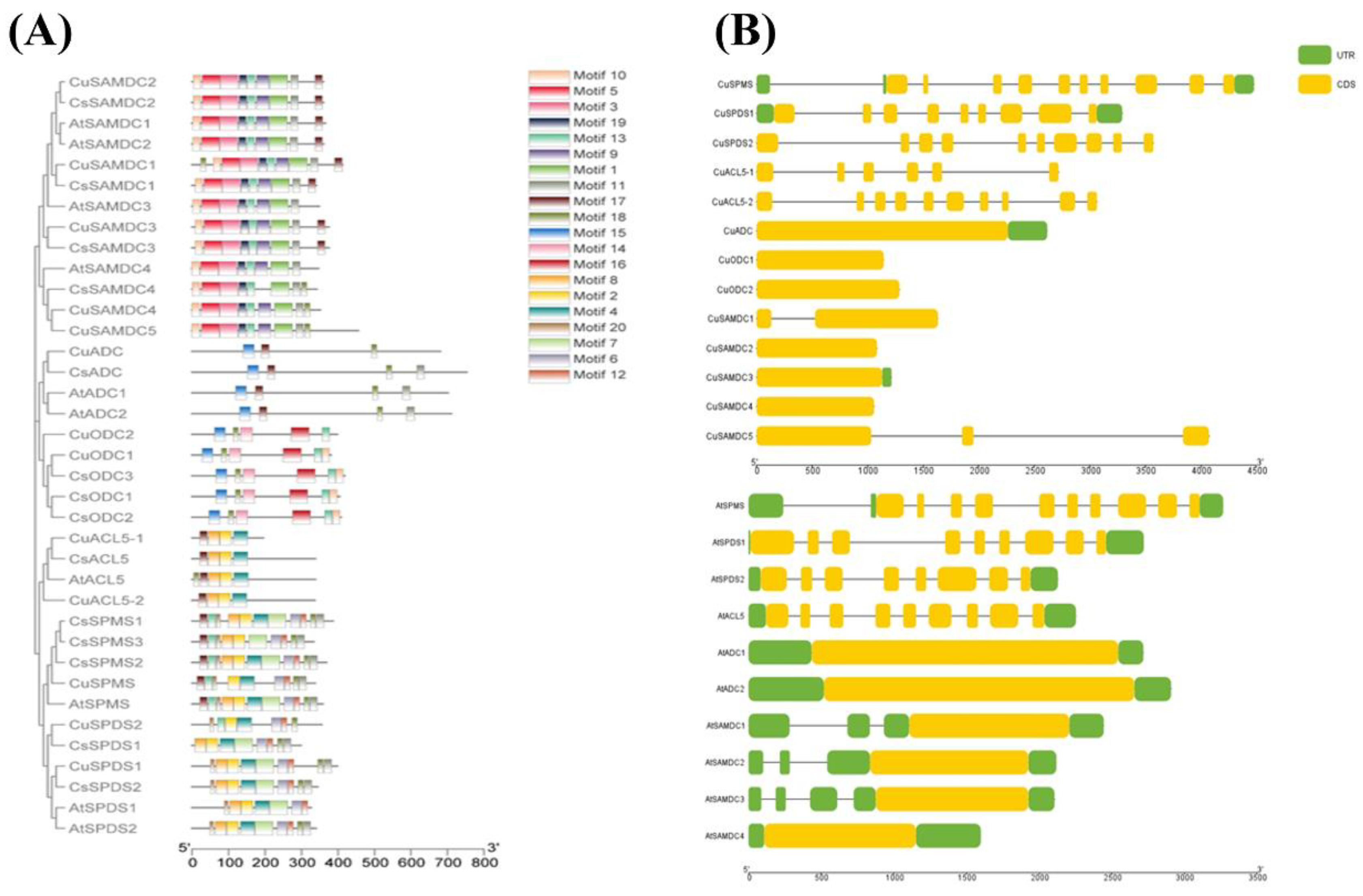
3.3. Identification of Conserved Motifs and Gene Structural Analysis
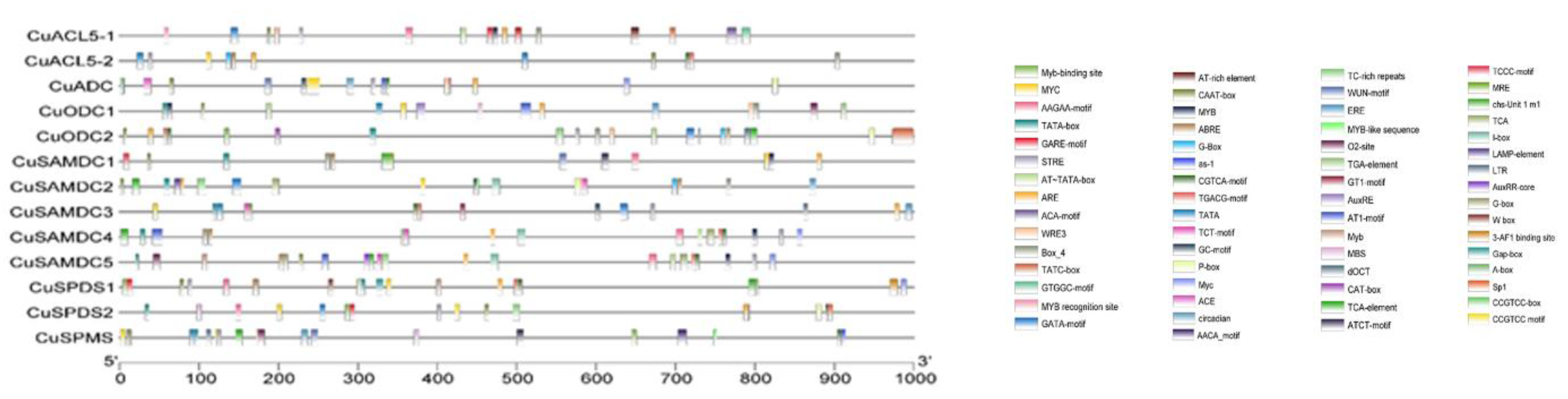
3.4. Cis-Elements Regulate the Transcriptional Activity of PA Biosynthetic Genes
3.5. Prediction Subcellular Localization Signals in PA Biosynthetic Genes of C. unshiu
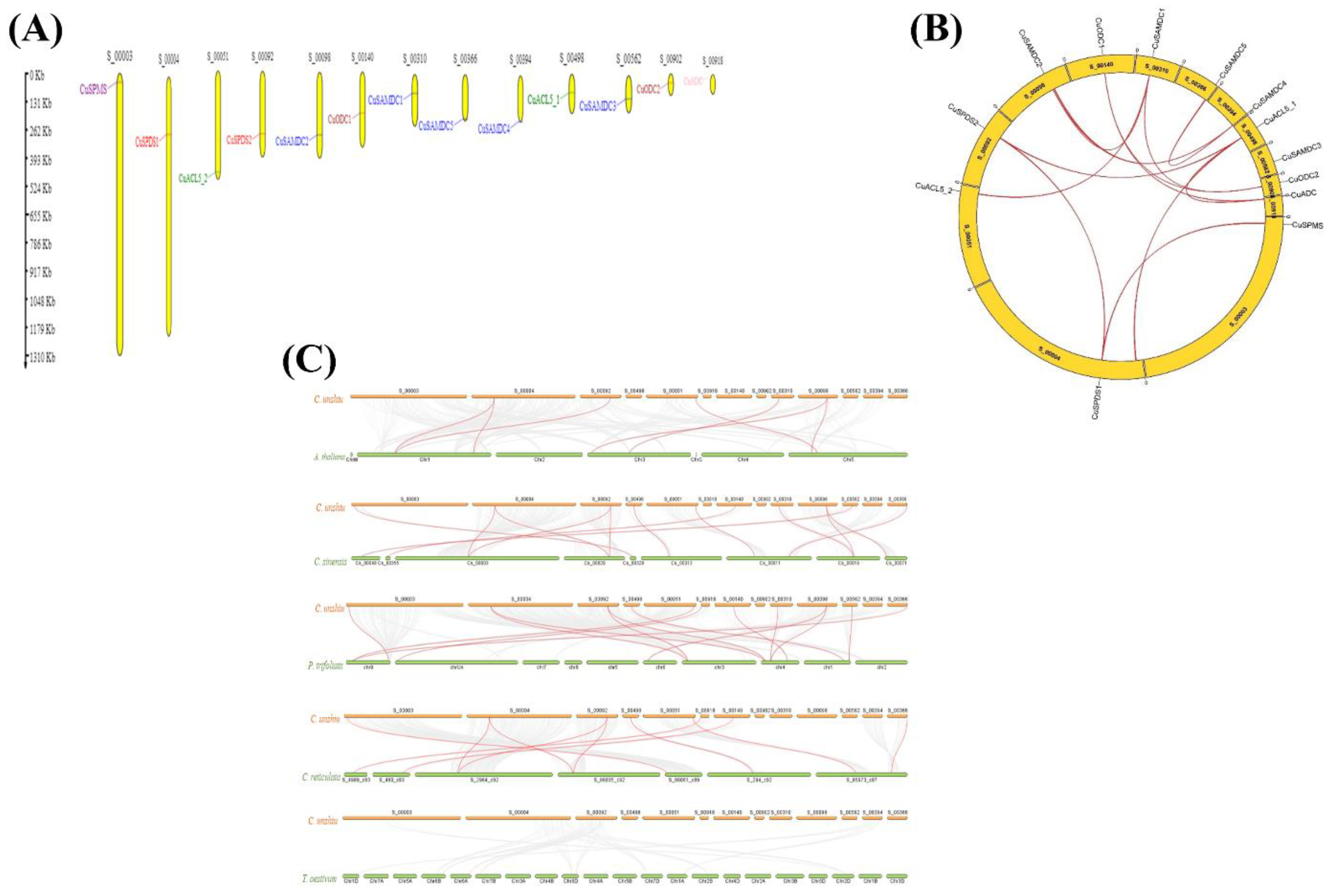
3.6. Gene Location and synteny Analysis in C. unshiu PA Biosynthetic Genes
3.7. Evaluation of C. unshiu PA Biosynthetic Genes Duplication Event
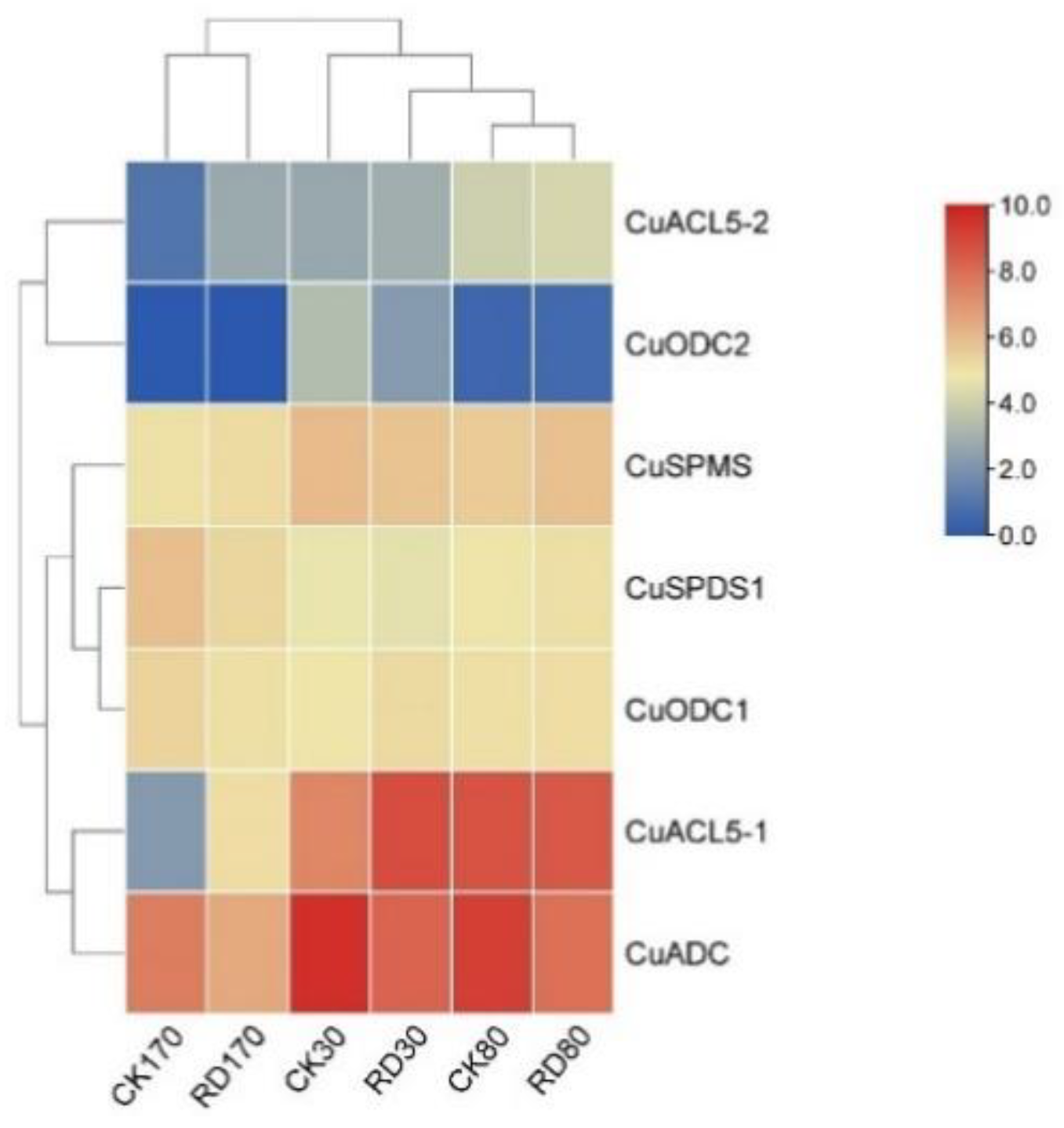
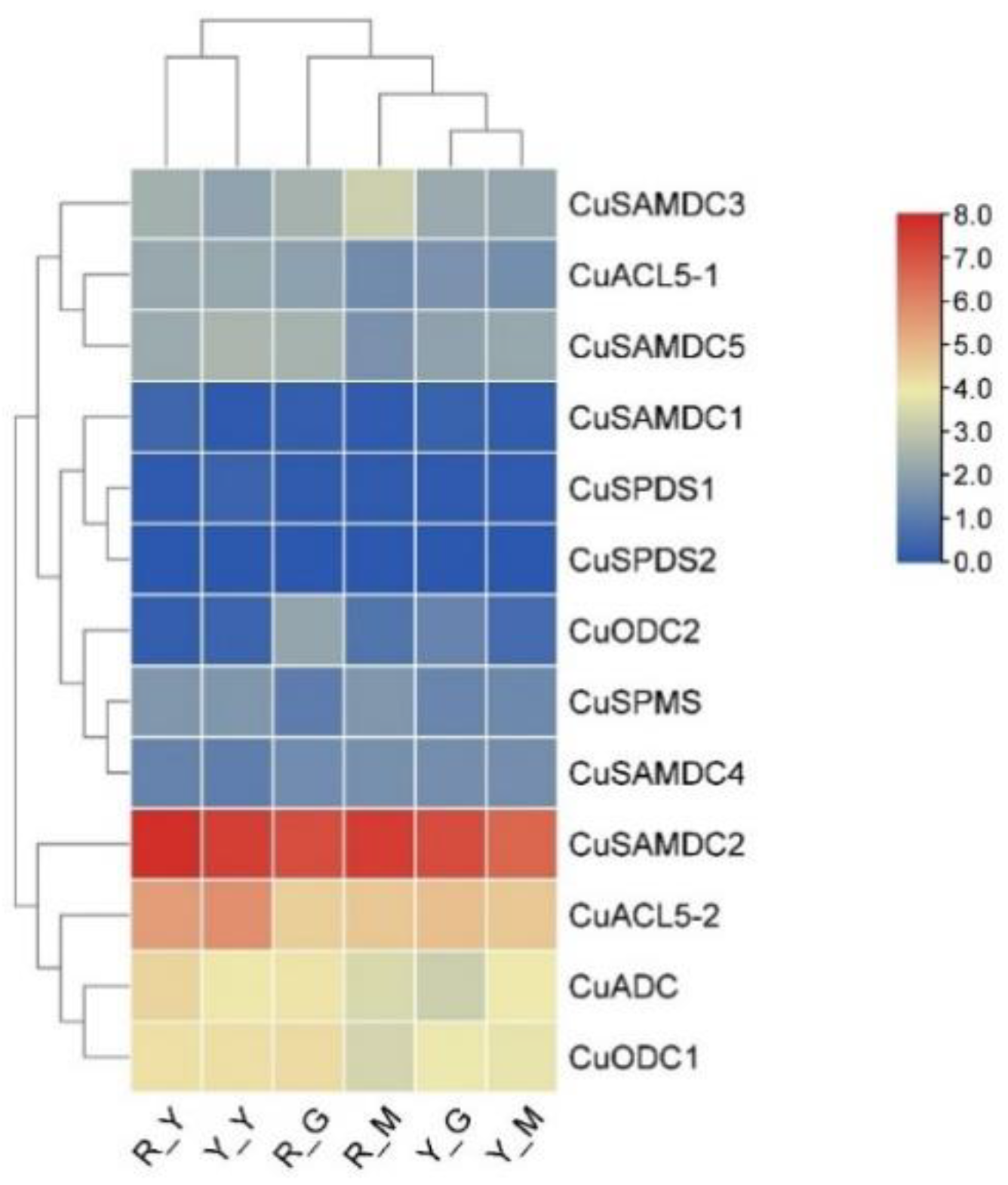
3.8. Expression of C. unshiu PA Biosynthetic Genes during Peel Roughing Disease
3.9. C. unshiu PA Biosynthetic Genes Showing Fluctuation in Expression during Seedling Etiolation in Two Citrus Hybrids
3.10. Differential Expression of PA Biosynthetic Genes in the Development of Light Drought-Induced Flowering Branches in C. unshiu
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aslıhan, Ö. The roles of polyamines in microorganisms. World J. Microbiol. Biotechnol. 2017, 33, 204. [Google Scholar]
- Singh, P.; Basu, S.; Kumar, G. Polyamines metabolism: A way ahead for abiotic stress tolerance in crop plants. In Biochemical, Physiological and Molecular Avenues for COMBATING Abiotic Stress Tolerance in Plants; Elsevier: Amsterdam, The Netherlands, 2018; pp. 39–55. [Google Scholar]
- Wickner, R.B. Herbert Tabor, 1918–2020: Polyamines, NIH, and the JBC. Proc. Natl. Acad. Sci. USA 2021, 118, e2023986118. [Google Scholar] [CrossRef]
- Hussain, M.; Iqbal, S.; Shafiq, M.; Balal, R.M.; Chater, J.; Kadyampakeni, D.; Alferez, F.; Sarkhosh, A.; Shahid, M.A. Silicon-induced hypoxia tolerance in citrus rootstocks associated with modulation in polyamine metabolism. Sci. Hortic. 2023, 318, 112118. [Google Scholar] [CrossRef]
- Wallace, H.M.; Fraser, A.V.; Hughes, A. A perspective of polyamine metabolism. Biochem. J. 2003, 376, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Seiler, N. A Guide to the Polyamines; University of Chicago Press: Chicago, IL, USA, 1999. [Google Scholar]
- Liu, J.-H.; Wang, W.; Wu, H.; Gong, X.; Moriguchi, T. Polyamines function in stress tolerance: From synthesis to regulation. Front. Plant Sci. 2015, 6, 827. [Google Scholar] [CrossRef]
- Michael, A.J. Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 2016, 473, 2315–2329. [Google Scholar] [CrossRef]
- Abouzari, A.; Solouki, M.; Golein, B.; Fakheri, B.A.; Sabouri, A. The change trend in physiological traits of 110 citrus accessions in response to cold stress. Bangladesh J. Bot. 2020, 49, 375–385. [Google Scholar] [CrossRef]
- Le, M.-L.; Sakai, K.; Mizunoe, Y.; Ozaki, Y.; Wakana, A. Evaluation of seedlings from 11 Citrus accessions for in vivo micrografting. Hortic. J. 2020, 89, 1–11. [Google Scholar] [CrossRef]
- Liu, Y.; Heying, E.; Tanumihardjo, S.A. History, global distribution, and nutritional importance of citrus fruits. Compr. Rev. Food Sci. Food Saf. 2012, 11, 530–545. [Google Scholar] [CrossRef]
- Gottwald, T.R. Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef]
- Zhou, C. The status of citrus Huanglongbing in China. Trop. Plant Pathol. 2020, 45, 279–284. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Xiao, L.-T.; Lu, X.-D.; Hu, J.-J.; Wu, S.; He, C.-Z.; Deng, X.-X. Changes in polyamine levels in Citrus sinensis Osb. cv. Valencia callus during somatic embryogenesis. J. Plant Physiol. Mol. Biol. 2005, 31, 275–280. [Google Scholar]
- Wu, X.-B.; Wang, J.; Liu, J.-H.; Deng, X.-X. Involvement of polyamine biosynthesis in somatic embryogenesis of Valencia sweet orange (Citrus sinensis) induced by glycerol. J. Plant Physiol. 2009, 166, 52–62. [Google Scholar] [CrossRef]
- Mendes, A.; Cidade, L.; Otoni, W.; Soares-Filho, W.; Costa, M. Role of auxins, polyamines and ethylene in root formation and growth in sweet orange. Biol. Plant. 2011, 55, 375–378. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, L.-R.; Xing, Q.-X.; Chen, J.-Z.; Zhu, H.-H. Exogenous polyamines influence root morphogenesis and arbuscular mycorrhizal development of Citrus limonia seedlings. Plant Growth Regul. 2010, 60, 27–33. [Google Scholar] [CrossRef]
- Anjum, M.A. Effect of NaCl concentrations in irrigation water on growth and polyamine metabolism in two citrus rootstocks with different levels of salinity tolerance. Acta Physiol. Plant. 2008, 30, 43–52. [Google Scholar] [CrossRef]
- Khoshbakht, D.; Asghari, M.; Haghighi, M. Influence of foliar application of polyamines on growth, gas-exchange characteristics, and chlorophyll fluorescence in Bakraii citrus under saline conditions. Photosynthetica 2018, 56, 731–742. [Google Scholar] [CrossRef]
- Hayat, F.; Li, J.; Liu, W.; Li, C.; Song, W.; Iqbal, S.; Khan, U.; Umer Javed, H.; Ahsan Altaf, M.; Tu, P. Influence of Citrus Rootstocks on Scion Growth, Hormone Levels, and Metabolites Profile of ‘Shatangju’Mandarin (Citrus reticulata Blanco). Horticulturae 2022, 8, 608. [Google Scholar] [CrossRef]
- Gentile, A.; Antognoni, F.; Iorio, R.A.; Distefano, G.; Las Casas, G.; La Malfa, S.; Serafini-Fracassini, D.; Del Duca, S. Polyamines and transglutaminase activity are involved in compatible and self-incompatible pollination of Citrus grandis. Amino Acids 2012, 42, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Sagee, O.; Lovatt, C.J. Putrescine Content Parallels Ammonia and Arginine Metabolism in Developing Flowers of theWashington’Navel Orange. J. Am. Soc. Hortic. Sci. 1991, 116, 280–285. [Google Scholar] [CrossRef]
- Hayat, F.; Li, J.; Iqbal, S.; Peng, Y.; Hong, L.; Balal, R.M.; Khan, M.N.; Nawaz, M.A.; Khan, U.; Farhan, M.A. A mini review of citrus rootstocks and their role in high-density orchards. Plants 2022, 11, 2876. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.K.; Dubey, A.; Srivastav, M.; Singh, A.; Sairam, R.; Pandey, R.; Dahuja, A.; Kaur, C. Effect of putrescine and paclobutrazol on growth, physiochemical parameters, and nutrient acquisition of salt-sensitive citrus rootstock Karna khatta (Citrus karna Raf.) under NaCl stress. J. Plant Growth Regul. 2011, 30, 301–311. [Google Scholar] [CrossRef]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Edgar, R. NCBI GEO: Mining tens of millions of expression profiles—Database and tools update. Nucleic Acids Res. 2007, 35 (Suppl. S1), D760–D765. [Google Scholar] [CrossRef]
- Edgar, R.; Barrett, T. NCBI GEO standards and services for microarray data. Nat. Biotechnol. 2006, 24, 1471–1472. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660, 289660. [Google Scholar]
- Kawahara, Y.; Endo, T.; Omura, M.; Teramoto, Y.; Itoh, T.; Fujii, H.; Shimada, T. Mikan Genome Database (MiGD): Integrated database of genome annotation, genomic diversity, and CAPS marker information for mandarin molecular breeding. Breed. Sci. 2020, 70, 200–211. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Horton, P.; Park, K.-J.; Obayashi, T.; Nakai, K. Protein subcellular localization prediction with WoLF PSORT. In Proceedings of the 4th Asia-Pacific Bioinformatics Conference, Taipei, Taiwan, 13–16 February 2006; World Scientific: Singapore, 2006; pp. 39–48. [Google Scholar]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Biłas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 127, 269–287. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef]
- Xing, Y.; Lee, C. Assessing the application of Ka/Ks ratio test to alternatively spliced exons. Bioinformatics 2005, 21, 3701–3703. [Google Scholar] [CrossRef] [PubMed]
- Parmley, J.L.; Hurst, L.D. How common are intragene windows with KA > KS owing to purifying selection on synonymous mutations? J. Mol. Evol. 2007, 64, 646–655. [Google Scholar] [CrossRef]
- Lu, X.-P.; Li, F.-F.; Xiong, J.; Cao, X.-J.; Ma, X.-C.; Zhang, Z.-M.; Cao, S.-Y.; Xie, S.-X. Transcriptome and metabolome analyses provide insights into the occurrence of peel roughing disorder on Satsuma Mandarin (Citrus unshiu Marc.) fruit. Front. Plant Sci. 2017, 8, 1907. [Google Scholar] [CrossRef]
- Bondarenko, V.S.; Gelfand, M.S. Evolution of the exon-intron structure in ciliate genomes. PLoS ONE 2016, 11, e0161476. [Google Scholar] [CrossRef]
- Koralewski, T.E.; Krutovsky, K.V. Evolution of exon-intron structure and alternative splicing. PLoS ONE 2011, 6, e18055. [Google Scholar] [CrossRef]
- Cai, Q.; Zhang, J.; Guo, C. Reviews of the physiological roles and molecular biology of polyamines in higher plants. J. Fujian Educ. Coll. 2006, 7, 118–124. [Google Scholar]
- Majumdar, R. Polyamine Metabolism in Arabidopsis: Transgenic Manipulation and Gene Expression. Doctoral Dissertations, University of New Hampshire, Durham, UK, 2011. [Google Scholar]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef]
- Taylor, J.S.; Raes, J. Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 2004, 38, 615–643. [Google Scholar] [CrossRef]
- Xie, T.; Zeng, L.; Chen, X.; Rong, H.; Wu, J.; Batley, J.; Jiang, J.; Wang, Y. Genome-wide analysis of the lateral organ boundaries domain gene family in Brassica napus. Genes 2020, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Liberles, D.A.; Teichmann, S.A.; Bahar, I.; Bastolla, U.; Bloom, J.; Bornberg Bauer, E.; Colwell, L.J.; De Koning, A.J.; Dokholyan, N.V.; Echave, J. The interface of protein structure, protein biophysics, and molecular evolution. Protein Sci. 2012, 21, 769–785. [Google Scholar] [CrossRef]
- Huang, B.; Jin, L.; Wang, P.; Wen, M.; Wu, S.; Xu, J. Methylome and Transcriptome Analysis of Flowering Branches Building of Citrus Plants Induced by Drought Stress. Gene 2023, 880, 147595. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Yang, H.; Ma, L.; Sun, N.; Yu, H.; Liu, T.; Gao, Y.; Gu, H.; Chen, Z.; Wada, M. A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol. 2003, 133, 1480–1493. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef]
- Ding, Z.; Weissmann, S.; Wang, M.; Du, B.; Huang, L.; Wang, L.; Tu, X.; Zhong, S.; Myers, C.; Brutnell, T.P. Identification of photosynthesis-associated C4 candidate genes through comparative leaf gradient transcriptome in multiple lineages of C3 and C4 species. PLoS ONE 2015, 10, e0140629. [Google Scholar] [CrossRef]


| Gene ID | Gene Name | Scaffold ID | Start | End | D | AA | MW (Kda) | pI |
|---|---|---|---|---|---|---|---|---|
| C_unshiu_00003:mRNA_4.1 | CuSPMS | C_unshiu_00003 | 40193 | 44670 | R | 338 | 38653.7 | 5.5 |
| C_unshiu_00004:mRNA_33.1 | CuSPDS1 | C_unshiu_00004 | 260305 | 263596 | R | 399 | 44011.5 | 5.2 |
| C_unshiu_00092:mRNA_48.1 | CuSPDS2 | C_unshiu_00092 | 340652 | 344223 | R | 356 | 40805.8 | 5.3 |
| C_unshiu_00498:mRNA_14.1 | CuACL5-1 | C_unshiu_00498 | 87851 | 90568 | R | 196 | 22151.4 | 5.6 |
| C_unshiu_00051:mRNA_69.1 | CuACL5-2 | C_unshiu_00051 | 560073 | 563136 | R | 337 | 37747.7 | 5.4 |
| C_unshiu_00918:mRNA_1.1 | CuADC | C_unshiu_00918 | 5010 | 7626 | F | 681 | 76854.2 | 5.1 |
| C_unshiu_00140:mRNA_18.1 | CuODC1 | C_unshiu_00140 | 217799 | 218944 | F | 381 | 41218 | 5.7 |
| C_unshiu_00902:mRNA_4.1 | CuODC2 | C_unshiu_00902 | 47979 | 49265 | F | 399 | 45316.1 | 7.2 |
| C_unshiu_00310:mRNA_10.1 | CuSAMDC1 | C_unshiu_00310 | 93929 | 95562 | F | 413 | 45414.5 | 5.2 |
| C_unshiu_00098:mRNA_50.1 | CuSAMDC2 | C_unshiu_00098 | 332348 | 333433 | F | 361 | 39849.1 | 5 |
| C_unshiu_00562:mRNA_14.1 | CuSAMDC3 | C_unshiu_00562 | 106938 | 108153 | R | 376 | 40852.4 | 5.5 |
| C_unshiu_00394:mRNA_35.1 | CuSAMDC4 | C_unshiu_00394 | 222794 | 223852 | F | 352 | 39289.6 | 5.2 |
| C_unshiu_00366:mRNA_33.1 | CuSAMDC5 | C_unshiu_00366 | 221007 | 225078 | F | 456 | 51360.6 | 5.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadiq, S.; Hussain, M.; Iqbal, S.; Shafiq, M.; Balal, R.M.; Seleiman, M.F.; Chater, J.; Shahid, M.A. Genome-Wide Identification and Characterization of the Biosynthesis of the Polyamine Gene Family in Citrus unshiu. Genes 2023, 14, 1527. https://doi.org/10.3390/genes14081527
Sadiq S, Hussain M, Iqbal S, Shafiq M, Balal RM, Seleiman MF, Chater J, Shahid MA. Genome-Wide Identification and Characterization of the Biosynthesis of the Polyamine Gene Family in Citrus unshiu. Genes. 2023; 14(8):1527. https://doi.org/10.3390/genes14081527
Chicago/Turabian StyleSadiq, Saleha, Mujahid Hussain, Shahid Iqbal, Muhammad Shafiq, Rashad Mukhtar Balal, Mahmoud F. Seleiman, John Chater, and Muhammad Adnan Shahid. 2023. "Genome-Wide Identification and Characterization of the Biosynthesis of the Polyamine Gene Family in Citrus unshiu" Genes 14, no. 8: 1527. https://doi.org/10.3390/genes14081527
APA StyleSadiq, S., Hussain, M., Iqbal, S., Shafiq, M., Balal, R. M., Seleiman, M. F., Chater, J., & Shahid, M. A. (2023). Genome-Wide Identification and Characterization of the Biosynthesis of the Polyamine Gene Family in Citrus unshiu. Genes, 14(8), 1527. https://doi.org/10.3390/genes14081527










