PromGER: Promoter Prediction Based on Graph Embedding and Ensemble Learning for Eukaryotic Sequence
Abstract
1. Introduction
2. Materials and Methods
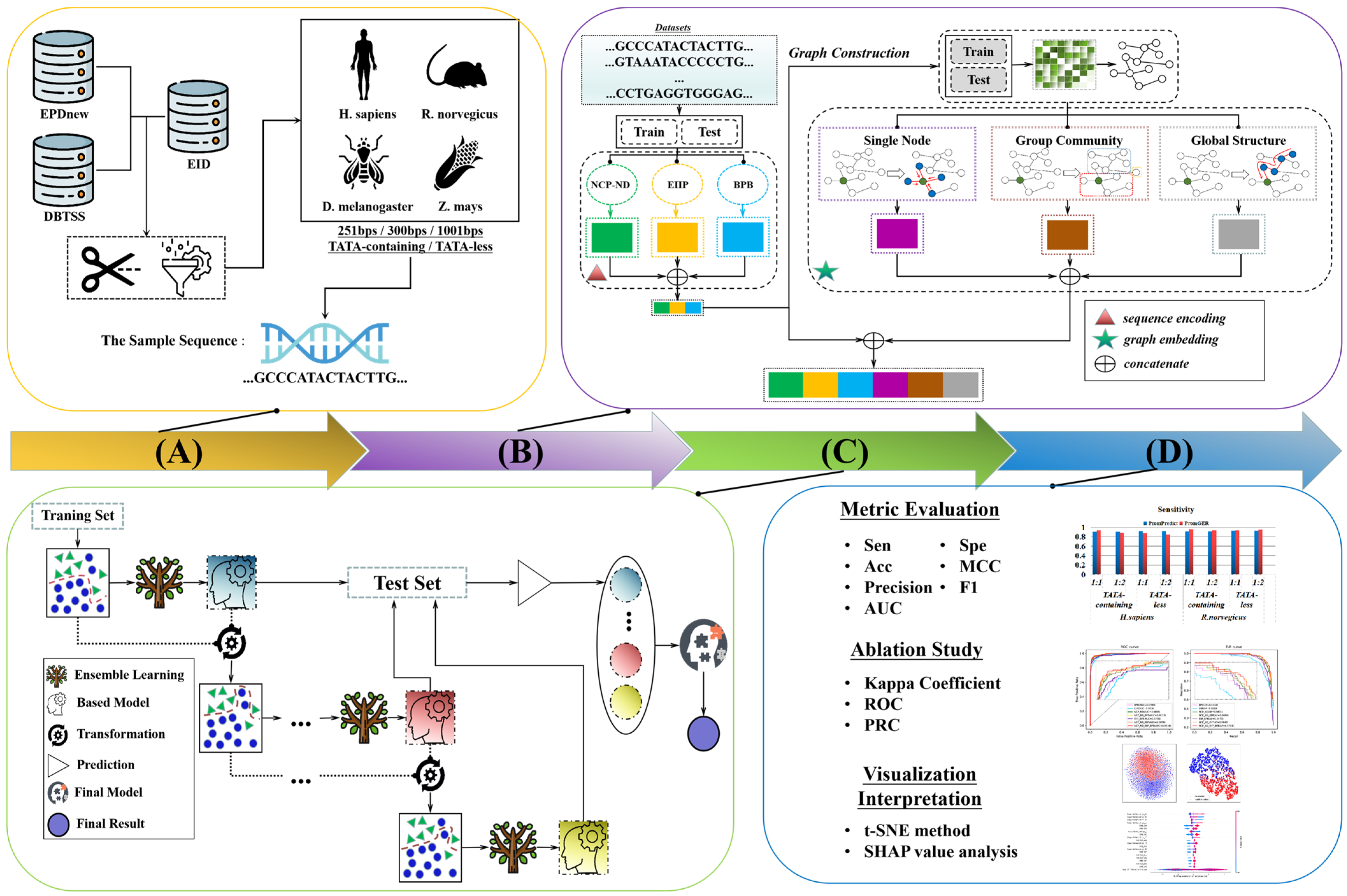
2.1. Overall Framework
2.2. Dataset Collecting
2.3. Sequence Feature-Encoding Method
2.3.1. Nucleotide Chemical Property (NCP) Feature Encoding
2.3.2. Nucleotide Density (ND) Feature Encoding
2.3.3. Electron–Ion Interaction Pseudopotential (EIIP) Feature Encoding
2.3.4. Bi-Profile Bayes (BPB) Feature Encoding
2.4. Graph Construction—Fast Linear Neighborhood Similarity Approach (FLNSA)
2.5. Graph-Embedding Feature-Encoding Method
2.5.1. Single Node—Node2vec
2.5.2. Group Community—SocDim
2.5.3. Global Structure—GraRep
2.6. Ensemble-Learning Strategy
3. Results
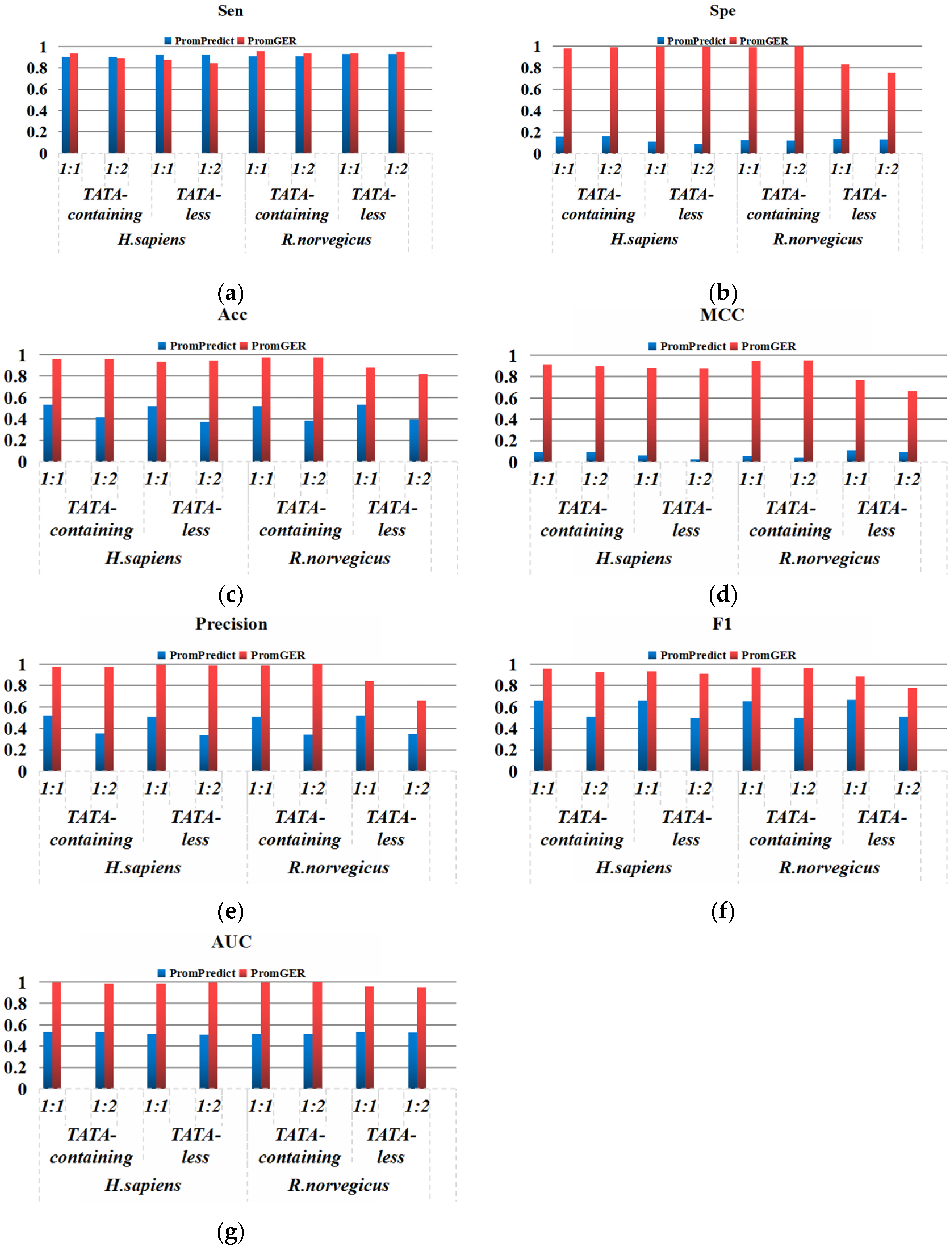
3.1. Performance Evaluation
3.2. Performance on Balanced Datasets
3.3. Performance on Imbalanced Datasets
3.4. Ablation Study of Different Feature Combinations
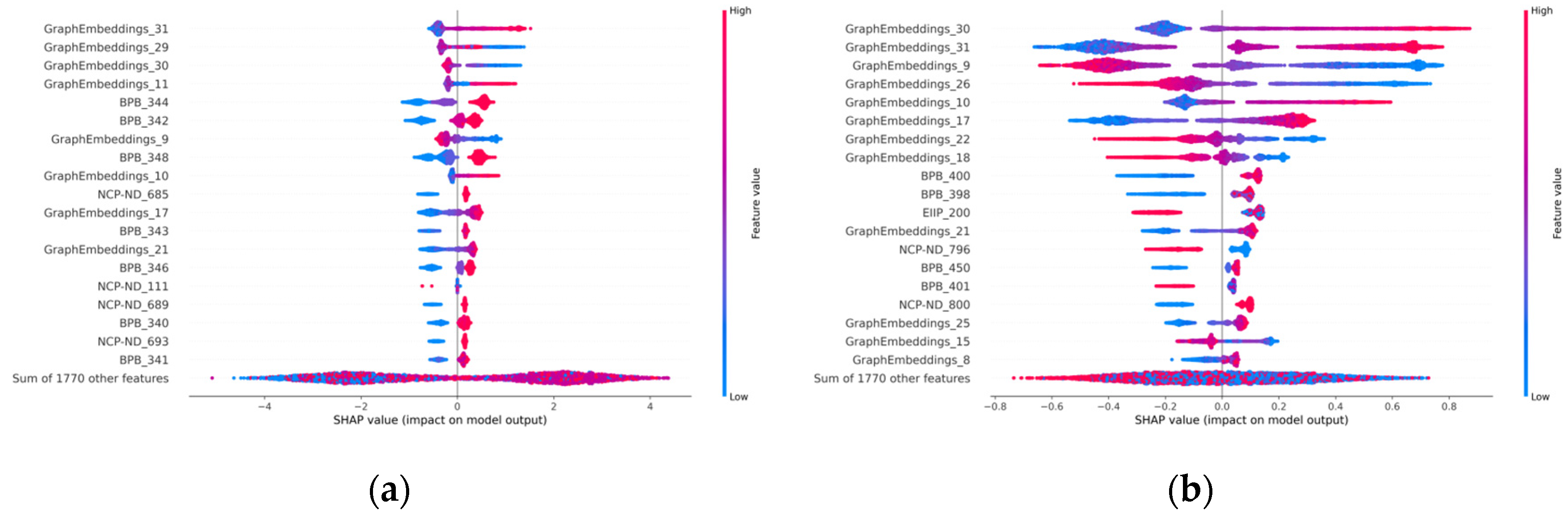
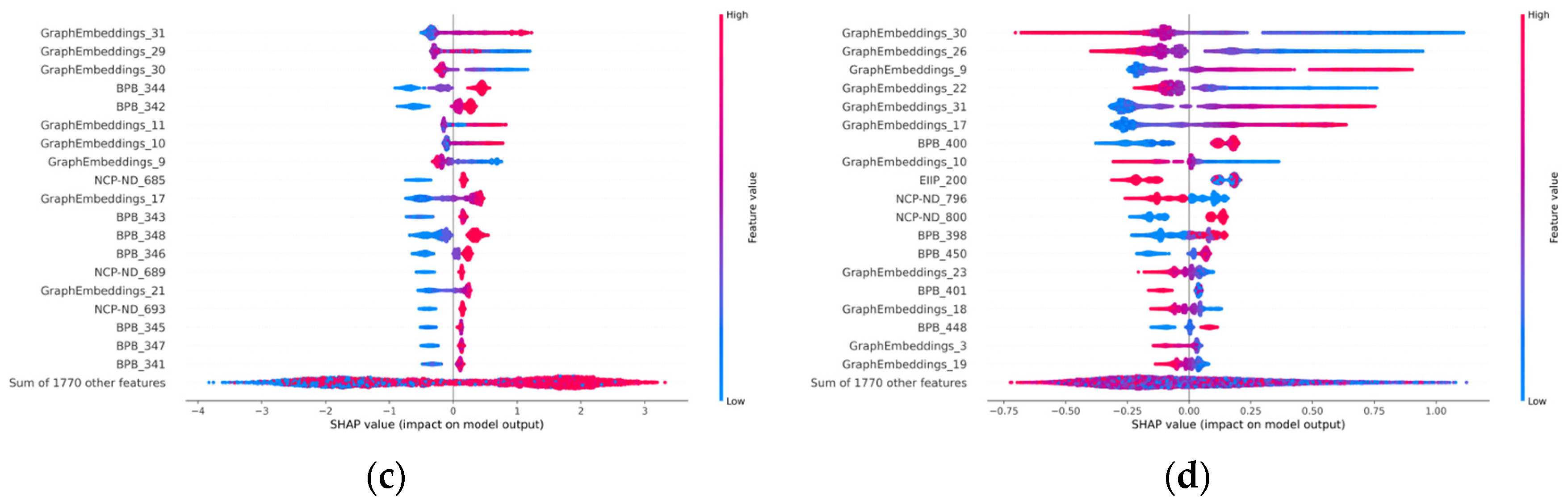
3.5. Model Visualization Interpretation
3.5.1. Visualization in Graph-Embedding Period
3.5.2. Visualization in Ensemble-Learning Periods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zeng, W.; Wang, F.; Ma, Y.; Liang, X.; Chen, P. Dysfunctional Mechanism of Liver Cancer Mediated by Transcription Factor and Non-coding RNA. Curr. Bioinform. 2019, 14, 100–107. [Google Scholar] [CrossRef]
- Azad, A.; Shahid, S.; Noman, N.; Lee, H. Prediction of plant promoters based on hexamers and random triplet pair analysis. Algorithms Mol. Biol. 2011, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Lin, Y.; Zou, Q. Transcription factors-DNA interactions in rice: Identification and verification. Brief. Bioinform. 2020, 21, 946–956. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Roberts, S. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005, 19, 2418–2423. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Sumazin, P.; Xuan, Z.; Zhang, M. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc. Natl. Acad. Sci. USA 2006, 103, 6275–6280. [Google Scholar] [CrossRef]
- Zhang, M. Computational analyses of eukaryotic promoters. BMC Bioinform. 2007, 8, S3. [Google Scholar] [CrossRef]
- Saif, I.; Kasmi, Y.; Allali, K.; Ennaji, M. Prediction of DNA methylation in the promoter of gene suppressor tumor. Gene 2018, 651, 166–173. [Google Scholar] [CrossRef]
- Kondapalli, M.; Galimudi, R.; Gundapaneni, K.; Padala, C.; Cingeetham, A.; Gantala, S.; Ali, A.; Shyamala, N.; Sahu, S.; Nallari, P.; et al. MMP 1 circulating levels and promoter polymorphism in risk prediction of coronary artery disease in asymptomatic first degree relatives. Gene 2016, 595, 115–120. [Google Scholar] [CrossRef]
- Gantala, S.; Kondapalli, M.; Kummari, R.; Padala, C.; Tupurani, M.; Kupsal, K.; Galimudi, R.; Gundapaneni, K.; Puranam, K.; Shyamala, N.; et al. Collagenase-1 (-1607 1G/2G), Gelatinase-A (-1306 C/T), Stromelysin-1 (-1171 5A/6A) functional promoter polymorphisms in risk prediction of type 2 diabetic nephropathy. Gene 2018, 673, 22–31. [Google Scholar] [CrossRef]
- Knudsen, S. Promoter2.0: For the recognition of PolII promoter sequences. Bioinformatics 1999, 15, 356–361. [Google Scholar] [CrossRef]
- Matsumine, H.; Yamamura, Y.; Hattori, N.; Kobayashi, T.; Kitada, T.; Yoritaka, A.; Mizuno, Y. A microdeletion of D6S305 in a family of autosomal recessive juvenile parkinsonism (PARK2). Genomics 1998, 49, 143–146. [Google Scholar] [CrossRef]
- Kim, J.W.; Zeller, K.I.; Wang, Y.; Jegga, A.G.; Aronow, B.J.; O’Donnell, K.A.; Dang, C.V. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 2004, 24, 5923–5936. [Google Scholar] [CrossRef]
- Dahl, J.A.; Collas, P. A rapid micro chromatin immunoprecipitation assay (microChIP). Nat. Protoc. 2008, 3, 1032–1045. [Google Scholar] [CrossRef]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child. Educ. Pract. Ed. 2013, 98, 236–238. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, Y.; Guo, B.; Ye, S.; Wu, Y.; Zhang, J. An improved promoter recognition model using convolutional neural network. In Proceedings of the 42nd Annual Computer Software and Applications Conference (COMPSAC 2018), Tokyo, Japan, 23–27 July 2018; pp. 471–476. [Google Scholar]
- Ponger, L.; Mouchiroud, D. CpGProD: Identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics 2002, 18, 631–633. [Google Scholar] [CrossRef]
- Wang, J.; Hannenhalli, S. A mammalian promoter model links cis elements to genetic networks. Biochem. Biophys. Res. Commun. 2006, 347, 166–177. [Google Scholar] [CrossRef]
- Scherf, M.; Klingenhoff, A.; Werner, T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: A novel context analysis approach. J. Mol. Biol. 2000, 297, 599–606. [Google Scholar] [CrossRef]
- Hutchinson, G.B. The prediction of vertebrate promoter regions using differential hexamer frequency analysis. Comput. Appl. Biosci. 1996, 12, 391–398. [Google Scholar] [CrossRef]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef]
- Chen, Q.K.; Hertz, G.Z.; Stormo, G.D. PromFD 1.0: A computer program that predicts eukaryotic pol II promoters using strings and IMD matrices. Comput. Appl. Biosci. 1997, 13, 29–35. [Google Scholar] [CrossRef]
- Down, T.A.; Hubbard, T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002, 12, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Aktar, U.; Jani, M.R.; Shatabda, S. iPro70-FMWin: Identifying Sigma70 promoters using multiple windowing and minimal features. Mol. Genet. Genom. 2019, 294, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; You, F. Optimization under uncertainty in the era of big data and deep learning: When machine learning meets mathematical programming. Comput. Chem. Eng. 2019, 125, 434–448. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.; Li, F.; Marquez-Lago, T.T.; Leier, A.; Revote, J.; Zhu, Y.; Powell, D.R.; Akutsu, T.; Webb, G.I.; et al. iLearn: An integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of DNA, RNA and protein sequence data. Brief. Bioinform. 2020, 21, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, F.; Marquez-Lago, T.T.; Leier, A.; Fan, C.; Kwoh, C.K.; Chou, K.C.; Song, J.; Jia, C. MULTiPly: A novel multi-layer predictor for discovering general and specific types of promoters. Bioinformatics 2019, 35, 2957–2965. [Google Scholar] [CrossRef]
- Lai, H.Y.; Zhang, Z.Y.; Su, Z.D.; Su, W.; Ding, H.; Chen, W.; Lin, H. iProEP: A Computational Predictor for Predicting Promoter. Mol. Ther. Nucleic Acids 2019, 17, 337–346. [Google Scholar] [CrossRef]
- Wang, X.; Xuan, Z.; Zhao, X.; Li, Y.; Zhang, M.Q. High-resolution human core-promoter prediction with CoreBoost_HM. Genome Res. 2009, 19, 266–275. [Google Scholar] [CrossRef]
- Zhao, X.; Xuan, Z.; Zhang, M.Q. Boosting with stumps for predicting transcription start sites. Genome Biol. 2007, 8, R17. [Google Scholar] [CrossRef]
- Prestridge, D.S. Predicting Pol II promoter sequences using transcription factor binding sites. J. Mol. Biol. 1995, 249, 923–932. [Google Scholar] [CrossRef]
- Davuluri, R.V.; Grosse, I.; Zhang, M.Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001, 29, 412–417. [Google Scholar] [CrossRef]
- Liu, B.; Yang, F.; Huang, D.S.; Chou, K.C. iPromoter-2L: A two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics 2018, 34, 33–40. [Google Scholar] [CrossRef]
- Bajic, V.B.; Seah, S.H.; Chong, A.; Zhang, G.; Koh, J.L.; Brusic, V. Dragon Promoter Finder: Recognition of vertebrate RNA polymerase II promoters. Bioinformatics 2002, 18, 198–199. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, J.; Wang, Y.; Li, S.; Jia, C.; Song, J.; Li, F. DeepTorrent: A deep learning-based approach for predicting DNA N4-methylcytosine sites. Brief. Bioinform. 2021, 22, bbaa124. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.; Li, F.; Wang, Y.; Smith, A.I.; Webb, G.I.; Akutsu, T.; Baggag, A.; Bensmail, H.; Song, J. Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Brief. Bioinform. 2020, 21, 1676–1696. [Google Scholar] [CrossRef]
- Li, F.; Chen, J.; Leier, A.; Marquez-Lago, T.; Liu, Q.; Wang, Y.; Revote, J.; Smith, A.I.; Akutsu, T.; Webb, G.I.; et al. DeepCleave: A deep learning predictor for caspase and matrix metalloprotease substrates and cleavage sites. Bioinformatics 2020, 36, 1057–1065. [Google Scholar] [CrossRef]
- Umarov, R.K.; Solovyev, V.V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS ONE 2017, 12, e0171410. [Google Scholar] [CrossRef]
- Oubounyt, M.; Louadi, Z.; Tayara, H.; Chong, K.T. DeePromoter: Robust Promoter Predictor Using Deep Learning. Front. Genet. 2019, 10, 286. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, F.; Xiang, D.; Akutsu, T.; Song, J.; Jia, C. Computational identification of eukaryotic promoters based on cascaded deep capsule neural networks. Brief. Bioinform. 2021, 22, bbaa299. [Google Scholar] [CrossRef]
- Sapoval, N.; Aghazadeh, A.; Nute, M.G.; Antunes, D.A.; Balaji, A.; Baraniuk, R.; Barberan, C.J.; Dannenfelser, R.; Dun, C.; Edrisi, M.; et al. Current progress and open challenges for applying deep learning across the biosciences. Nat. Commun. 2022, 13, 1728. [Google Scholar] [CrossRef]
- Zhang, X.M.; Liang, L.; Liu, L.; Tang, M.J. Graph Neural Networks and Their Current Applications in Bioinformatics. Front. Genet. 2021, 12, 690049. [Google Scholar] [CrossRef]
- Li, H.; Shi, L.; Gao, W.; Zhang, Z.; Zhang, L.; Zhao, Y.; Wang, G. dPromoter-XGBoost: Detecting promoters and strength by combining multiple descriptors and feature selection using XGBoost. Methods 2022, 204, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, J.; Ge, Z.; Wen, Y.; Yue, Y.; Hayashida, M.; Baggag, A.; Bensmail, H.; Song, J. Computational prediction and interpretation of both general and specific types of promoters in Escherichia coli by exploiting a stacked ensemble-learning framework. Brief. Bioinform. 2021, 22, 2126–2140. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, R.; Huang, L.; Yang, S.; Hu, X.; He, K. m6AGE: A Predictor for N6-Methyladenosine Sites Identification Utilizing Sequence Characteristics and Graph Embedding-Based Geometrical Information. Front. Genet. 2021, 12, 670852. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef] [PubMed]
- Yella, V.R.; Kumar, A.; Bansal, M. Identification of putative promoters in 48 eukaryotic genomes on the basis of DNA free energy. Sci. Rep. 2018, 8, 4520. [Google Scholar] [CrossRef]
- Lv, H.; Dao, F.Y.; Zhang, D.; Guan, Z.X.; Yang, H.; Su, W.; Liu, M.L.; Ding, H.; Chen, W.; Lin, H. iDNA-MS: An Integrated Computational Tool for Detecting DNA Modification Sites in Multiple Genomes. iScience 2020, 23, 100991. [Google Scholar] [CrossRef]
- Zhang, M.; Jia, C.; Li, F.; Li, C.; Zhu, Y.; Akutsu, T.; Webb, G.I.; Zou, Q.; Coin, L.; Song, J. Critical assessment of computational tools for prokaryotic and eukaryotic promoter prediction. Brief. Bioinform. 2022, 23, bbab551. [Google Scholar] [CrossRef]
- Dreos, R.; Ambrosini, G.; Périer, R.C.; Bucher, P. The Eukaryotic Promoter Database: Expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015, 43, 92–96. [Google Scholar] [CrossRef]
- Yamashita, R.; Sugano, S.; Suzuki, Y.; Nakai, K. DBTSS: DataBase of Transcriptional Start Sites progress report in 2012. Nucleic Acids Res. 2012, 40, 150–154. [Google Scholar] [CrossRef]
- Shepelev, V.; Fedorov, A. Advances in the Exon-Intron Database (EID). Brief. Bioinform. 2006, 7, 178–185. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Zou, Q.; Lin, G.; Jiang, X.; Liu, X.; Zeng, X. Sequence clustering in bioinformatics: An empirical study. Brief. Bioinform. 2020, 21, 1–10. [Google Scholar] [CrossRef]
- Xu, M.; Gonzalez-Hurtado, E.; Martinez, E. Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription. Biochim. Biophys. Acta 2016, 1859, 553–563. [Google Scholar] [CrossRef]
- Jeong, B.S.; Golam, A.T.; Rokeya, M.; Jeon, S.; Lim, C.G.; Choi, H.J. Codon-based encoding for DNA sequence analysis. Methods 2014, 67, 373–379. [Google Scholar] [CrossRef]
- Bari, A.; Reaz, M.; Choi, H.; Jeong, B. DNA Encoding for Splice Site Prediction in Large DNA Sequence. In Proceedings of the Database Systems for Advanced Applications, (DASFAA 2013), Wuhan, China, 22–25 April 2013; pp. 46–58. [Google Scholar]
- Nair, A.S.; Sreenadhan, S.P. A coding measure scheme employing electron-ion interaction pseudopotential (EIIP). Bioinformation 2006, 1, 197–202. [Google Scholar]
- Shao, J.; Xu, D.; Tsai, S.N.; Wang, Y.; Ngai, S.M. Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PLoS ONE 2009, 4, e4920. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, G.; Zhou, S.; Niu, Y. LncRNA-miRNA interaction prediction through sequence-derived linear neighborhood propagation method with information combination. BMC Genom. 2019, 20, 946. [Google Scholar] [CrossRef]
- Goyal, P.; Ferrara, E. Graph embedding techniques, applications, and performance: A survey. Knowl.-Based Syst. 2018, 151, 78–94. [Google Scholar] [CrossRef]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (SIGKDD 2016), San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Tang, L.; Liu, H. Relational learning via latent social dimensions. In Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (SIGKDD 2009), Paris, France, 28 June–1 July 2009; pp. 817–826. [Google Scholar]
- Cao, S.; Lu, W.; Xu, Q. Grarep: Learning graph representations with global structural information. In Proceedings of the 24th ACM International on Conference on Information and Knowledge Management (CIKM 2015), Melbourne, Australia, 19–23 October 2015; pp. 891–900. [Google Scholar]
- Huang, G.; Wu, L.; Ma, X.; Zhang, W.; Fan, J.; Yu, X.; Zeng, W.; Zhou, H. Evaluation of CatBoost method for prediction of reference evapotranspiration in humid regions. J. Hydrol. 2019, 574, 1029–1041. [Google Scholar] [CrossRef]



| Species | TATA | Number of Promoters | Number of Non-Promoters | Location | Length |
|---|---|---|---|---|---|
| Homo sapiens | TATA-containing | 229 | 229 | [−200, +50] | 251 |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| TATA-less | 1328 | 1328 | [−200, +50] | 251 | |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| Rattus norvegicus | TATA-containing | 456 | 456 | [−200, +50] | 251 |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| TATA-less | 3834 | 3834 | [−200, +50] | 251 | |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| Drosophila melanogaster | TATA-containing | 207 | 207 | [−249, +50] | 300 |
| TATA-less | 1153 | 1153 | [−249, +50] | 300 | |
| Zea mays | TATA-containing | 3051 | 3051 | [−249, +50] | 300 |
| TATA-less | 5210 | 5210 | [−249, +50] | 300 |
| Species | TATA | Number of Promoters | Number of Non-Promoters | Location | Length |
|---|---|---|---|---|---|
| Homo sapiens | TATA-containing | 229 | 458 | [−200, +50] | 251 |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| TATA-less | 1328 | 2656 | [−200, +50] | 251 | |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| Rattus norvegicus | TATA-containing | 456 | 912 | [−200, +50] | 251 |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| TATA-less | 3834 | 7668 | [−200, +50] | 251 | |
| [−249, +50] | 300 | ||||
| [−500, +500] | 1001 | ||||
| Drosophila melanogaster | TATA-containing | 207 | 1035 | [−249, +50] | 300 |
| TATA-less | 1153 | 5765 | [−249, +50] | 300 | |
| Zea mays | TATA-containing | 610 | 3050 | [−249, +50] | 300 |
| TATA-less | 2762 | 13,810 | [−249, +50] | 300 |
| Datasets | Models | Sen | Spe | Acc | MCC | Pre | F1 | AUC | |
|---|---|---|---|---|---|---|---|---|---|
| H. sapiens (251 bps) | TATA- containing | CNNProm | 0.8559 | 0.9825 | 0.9192 | 0.8452 | 0.9800 | 0.9138 | 0.9250 |
| NNPP2.2 | 0.9520 | 0.4279 | 0.6900 | 0.4461 | 0.6246 | 0.7543 | 0.6900 | ||
| FProm | 0.4978 | 0.9738 | 0.7358 | 0.5363 | 0.9500 | 0.6533 | 0.7358 | ||
| PromGER | 0.9565 | 0.9782 | 0.9673 | 0.9350 | 0.9772 | 0.9670 | 0.9952 | ||
| TATA- less | CNNProm | 0.8504 | 0.7995 | 0.8164 | 0.6212 | 0.6791 | 0.7551 | 0.8984 | |
| NNPP2.2 | 0.7417 | 0.3742 | 0.5580 | 0.1247 | 0.5424 | 0.6266 | 0.5580 | ||
| FProm | 0.4834 | 0.9571 | 0.7203 | 0.5002 | 0.9185 | 0.6334 | 0.7203 | ||
| PromGER | 0.8646 | 0.9849 | 0.9248 | 0.8558 | 0.9829 | 0.9199 | 0.9844 | ||
| H. sapiens (300 bps) | TATA- containing | iProEP | 0.7249 | 0.9476 | 0.8362 | 0.6898 | 0.9326 | 0.8157 | 0.9416 |
| Depicter | 0.9607 | 0.8996 | 0.9301 | 0.8619 | 0.9053 | 0.9322 | 0.9715 | ||
| DeePromoter | 0.9520 | 0.4279 | 0.6900 | 0.4461 | 0.6246 | 0.7543 | 0.6900 | ||
| PromGER | 0.9113 | 0.9757 | 0.9424 | 0.8908 | 0.9761 | 0.9425 | 0.9975 | ||
| TATA- less | iProEP | 0.6190 | 0.9262 | 0.7726 | 0.5729 | 0.8935 | 0.7313 | 0.8589 | |
| Depicter | 0.7869 | 0.8253 | 0.8061 | 0.6127 | 0.8183 | 0.8023 | 0.8446 | ||
| DeePromoter | 0.9149 | 0.1453 | 0.5301 | 0.0943 | 0.5170 | 0.6607 | 0.5301 | ||
| PromGER | 0.8905 | 0.9849 | 0.9377 | 0.8793 | 0.9833 | 0.9346 | 0.9832 | ||
| R. norvegicus (251 bps) | TATA- containing | CNNProm | 0.9298 | 0.9649 | 0.9474 | 0.8953 | 0.9636 | 0.9464 | 0.9596 |
| NNPP2.2 | 0.9452 | 0.4232 | 0.6842 | 0.4319 | 0.6210 | 0.7496 | 0.6842 | ||
| PromGER | 0.9350 | 0.9670 | 0.9560 | 0.9123 | 0.9662 | 0.9555 | 0.9923 | ||
| TATA- less | CNNProm | 0.8263 | 0.8905 | 0.8584 | 0.7182 | 0.8829 | 0.8537 | 0.8883 | |
| NNPP2.2 | 0.7433 | 0.4520 | 0.5977 | 0.2042 | 0.5756 | 0.6488 | 0.5977 | ||
| PromGER | 0.9425 | 0.7819 | 0.8596 | 0.7281 | 0.8112 | 0.8697 | 0.9488 | ||
| R. norvegicus (300 bps) | TATA- containing | Depicter | 0.9846 | 0.7522 | 0.8684 | 0.7576 | 0.7989 | 0.8821 | 0.9786 |
| DeePromoter | 0.8158 | 0.4781 | 0.6469 | 0.3122 | 0.6098 | 0.6979 | 0.6469 | ||
| PromGER | 0.9340 | 0.9450 | 0.9395 | 0.8791 | 0.9444 | 0.9392 | 0.9841 | ||
| TATA- less | Depicter | 0.9726 | 0.7467 | 0.8597 | 0.7384 | 0.7934 | 0.8739 | 0.8839 | |
| DeePromoter | 0.8192 | 0.2433 | 0.5313 | 0.0766 | 0.5199 | 0.6361 | 0.5313 | ||
| PromGER | 0.9516 | 0.7806 | 0.8661 | 0.7433 | 0.8127 | 0.8767 | 0.9485 | ||
| Datasets | Models | Sen | Spe | Acc | MCC | Pre | F1 | AUC | |
|---|---|---|---|---|---|---|---|---|---|
| H. sapiens (251 bps) | TATA- containing | CNNProm | 0.8518 | 0.8686 | 0.8602 | 0.7205 | 0.8664 | 0.8590 | 0.8979 |
| NNPP2.2 | 0.9520 | 0.4279 | 0.6026 | 0.3903 | 0.4542 | 0.6150 | 0.6900 | ||
| FProm | 0.4978 | 0.8996 | 0.7656 | 0.4432 | 0.7125 | 0.5861 | 0.6987 | ||
| PromGER | 0.8666 | 0.9780 | 0.9411 | 0.8661 | 0.9512 | 0.9069 | 0.9736 | ||
| TATA- less | CNNProm | 0.8124 | 0.5549 | 0.7089 | 0.4391 | 0.6597 | 0.7477 | 0.7089 | |
| NNPP2.2 | 0.7417 | 0.3852 | 0.5040 | 0.1260 | 0.3762 | 0.4992 | 0.5634 | ||
| FProm | 0.4834 | 0.8991 | 0.7605 | 0.4295 | 0.7055 | 0.5737 | 0.6913 | ||
| PromGER | 0.8679 | 0.9962 | 0.9535 | 0.8961 | 0.9913 | 0.9255 | 0.9932 | ||
| H. sapiens (300 bps) | TATA- containing | iProEP | 0.7249 | 0.9585 | 0.8806 | 0.7263 | 0.8973 | 0.8019 | 0.9495 |
| Depicter | 0.9607 | 0.9170 | 0.9316 | 0.8544 | 0.8527 | 0.9035 | 0.9438 | ||
| DeePromoter | 0.9520 | 0.4760 | 0.6346 | 0.4279 | 0.4760 | 0.6346 | 0.7140 | ||
| PromGER | 0.8666 | 0.9670 | 0.9338 | 0.8490 | 0.9285 | 0.8965 | 0.9755 | ||
| TATA- less | iProEP | 0.6190 | 0.9326 | 0.8281 | 0.5995 | 0.8212 | 0.7059 | 0.8576 | |
| Depicter | 0.7869 | 0.7952 | 0.7924 | 0.5604 | 0.6576 | 0.7165 | 0.7640 | ||
| DeePromoter | 0.9149 | 0.1224 | 0.3865 | 0.0562 | 0.3426 | 0.4986 | 0.5186 | ||
| PromGER | 0.8754 | 0.9962 | 0.9560 | 0.9017 | 0.9914 | 0.9298 | 0.9871 | ||
| R. norvegicus (251 bps) | TATA- containing | CNNProm | 0.9578 | 0.9641 | 0.9610 | 0.9220 | 0.9639 | 0.9608 | 0.9757 |
| NNPP2.2 | 0.9452 | 0.4243 | 0.5980 | 0.3797 | 0.4508 | 0.6105 | 0.6848 | ||
| PromGER | 0.9350 | 0.9945 | 0.9780 | 0.9505 | 0.9885 | 0.9662 | 0.9972 | ||
| TATA- less | CNNProm | 0.6934 | 0.5415 | 0.6174 | 0.2376 | 0.5519 | 0.6444 | 0.6174 | |
| NNPP2.2 | 0.7433 | 0.4384 | 0.5401 | 0.1768 | 0.3983 | 0.5187 | 0.5909 | ||
| PromGER | 0.9451 | 0.6784 | 0.7672 | 0.5888 | 0.5949 | 0.7302 | 0.9290 | ||
| R. norvegicus (300 bps) | TATA- containing | Depicter | 0.9846 | 0.8169 | 0.8728 | 0.7595 | 0.7289 | 0.8377 | 0.9267 |
| DeePromoter | 0.8158 | 0.4386 | 0.5643 | 0.2508 | 0.4208 | 0.5552 | 0.6272 | ||
| PromGER | 0.9560 | 0.9835 | 0.9743 | 0.9421 | 0.9666 | 0.9613 | 0.9952 | ||
| TATA- less | Depicter | 0.9726 | 0.7851 | 0.8476 | 0.7159 | 0.6935 | 0.8097 | 0.8480 | |
| DeePromoter | 0.8192 | 0.2237 | 0.4222 | 0.0497 | 0.3454 | 0.4859 | 0.5215 | ||
| PromGER | 0.9373 | 0.9680 | 0.9578 | 0.9050 | 0.9361 | 0.9367 | 0.9902 | ||
| Feature Combinations | Kappa | |||||
|---|---|---|---|---|---|---|
| Promoters:Non-Promoters = 1:1 | Promoters:Non-Promoters = 1:5 | |||||
| NCP_ND | EIIP | BPB | TATA-Containing | TATA-Less | TATA-Containing | TATA-Less |
| √ | 0.7317 | 0.8913 | 0.8189 | 0.8699 | ||
| √ | 0.8048 | 0.8608 | 0.8043 | 0.8027 | ||
| √ | 0.8495 | 0.8826 | 0.8031 | 0.8207 | ||
| √ | √ | 0.7804 | 0.8869 | 0.7786 | 0.8801 | |
| √ | √ | 0.8516 | 0.8913 | 0.7993 | 0.8421 | |
| √ | √ | 0.7804 | 0.8956 | 0.8031 | 0.8572 | |
| √ | √ | √ | 0.8536 | 0.9130 | 0.8343 | 0.8833 |
| Feature Combinations | Kappa | |||||
|---|---|---|---|---|---|---|
| Promoters:Non-Promoters = 1:1 | Promoters:Non-Promoters = 1:5 | |||||
| Node2vec | SocDim | GraRep | TATA-Containing | TATA-Less | TATA-Containing | TATA-Less |
| √ | 0.8591 | 0.8958 | 0.8415 | 0.8908 | ||
| √ | 0.8390 | 0.9079 | 0.8213 | 0.8838 | ||
| √ | 0.8835 | 0.9283 | 0.8507 | 0.8627 | ||
| √ | √ | 0.9193 | 0.9097 | 0.8498 | 0.8919 | |
| √ | √ | 0.8611 | 0.9205 | 0.8451 | 0.8776 | |
| √ | √ | 0.8730 | 0.9113 | 0.8364 | 0.8878 | |
| √ | √ | √ | 0.9238 | 0.9439 | 0.8512 | 0.8968 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Tai, S.; Zhang, S.; Sheng, N.; Xie, X. PromGER: Promoter Prediction Based on Graph Embedding and Ensemble Learning for Eukaryotic Sequence. Genes 2023, 14, 1441. https://doi.org/10.3390/genes14071441
Wang Y, Tai S, Zhang S, Sheng N, Xie X. PromGER: Promoter Prediction Based on Graph Embedding and Ensemble Learning for Eukaryotic Sequence. Genes. 2023; 14(7):1441. https://doi.org/10.3390/genes14071441
Chicago/Turabian StyleWang, Yan, Shiwen Tai, Shuangquan Zhang, Nan Sheng, and Xuping Xie. 2023. "PromGER: Promoter Prediction Based on Graph Embedding and Ensemble Learning for Eukaryotic Sequence" Genes 14, no. 7: 1441. https://doi.org/10.3390/genes14071441
APA StyleWang, Y., Tai, S., Zhang, S., Sheng, N., & Xie, X. (2023). PromGER: Promoter Prediction Based on Graph Embedding and Ensemble Learning for Eukaryotic Sequence. Genes, 14(7), 1441. https://doi.org/10.3390/genes14071441






