Escherichia albertii as a Potential Enteropathogen in the Light of Epidemiological and Genomic Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples and Bacterial Cultures
2.2. PCR Detection of E. albertii, Enteropathogenic E. coli (EPEC), Enterohemorrhagic E. coli (EHEC) and Avian Pathogenic E. coli (APEC) Strains
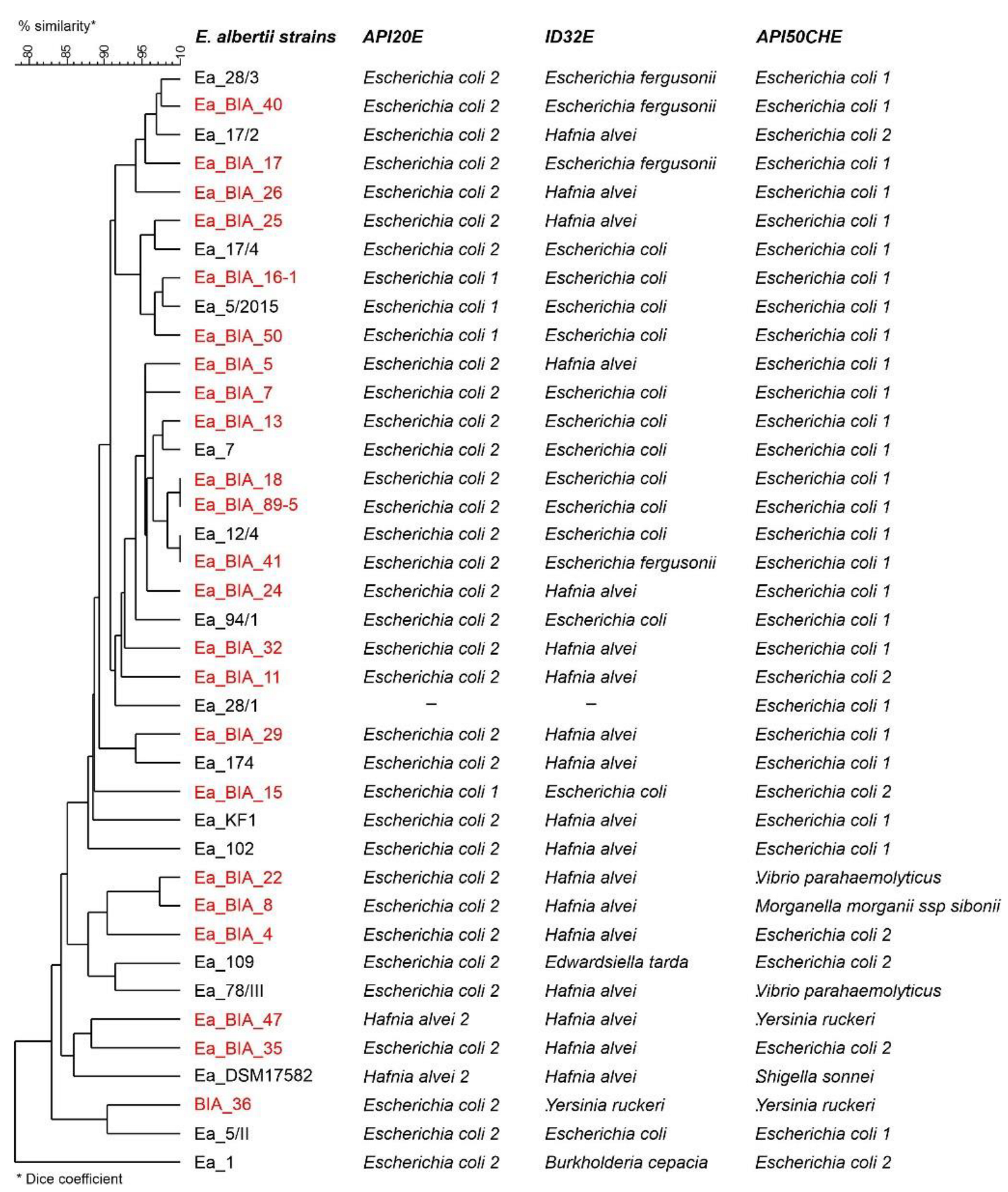
2.3. Phenotypic Features of E. albertii
Biochemical Profiles
2.4. Antimicrobial Susceptibility Patterns
- Determination of Genetic Relatedness
2.5. Serotyping
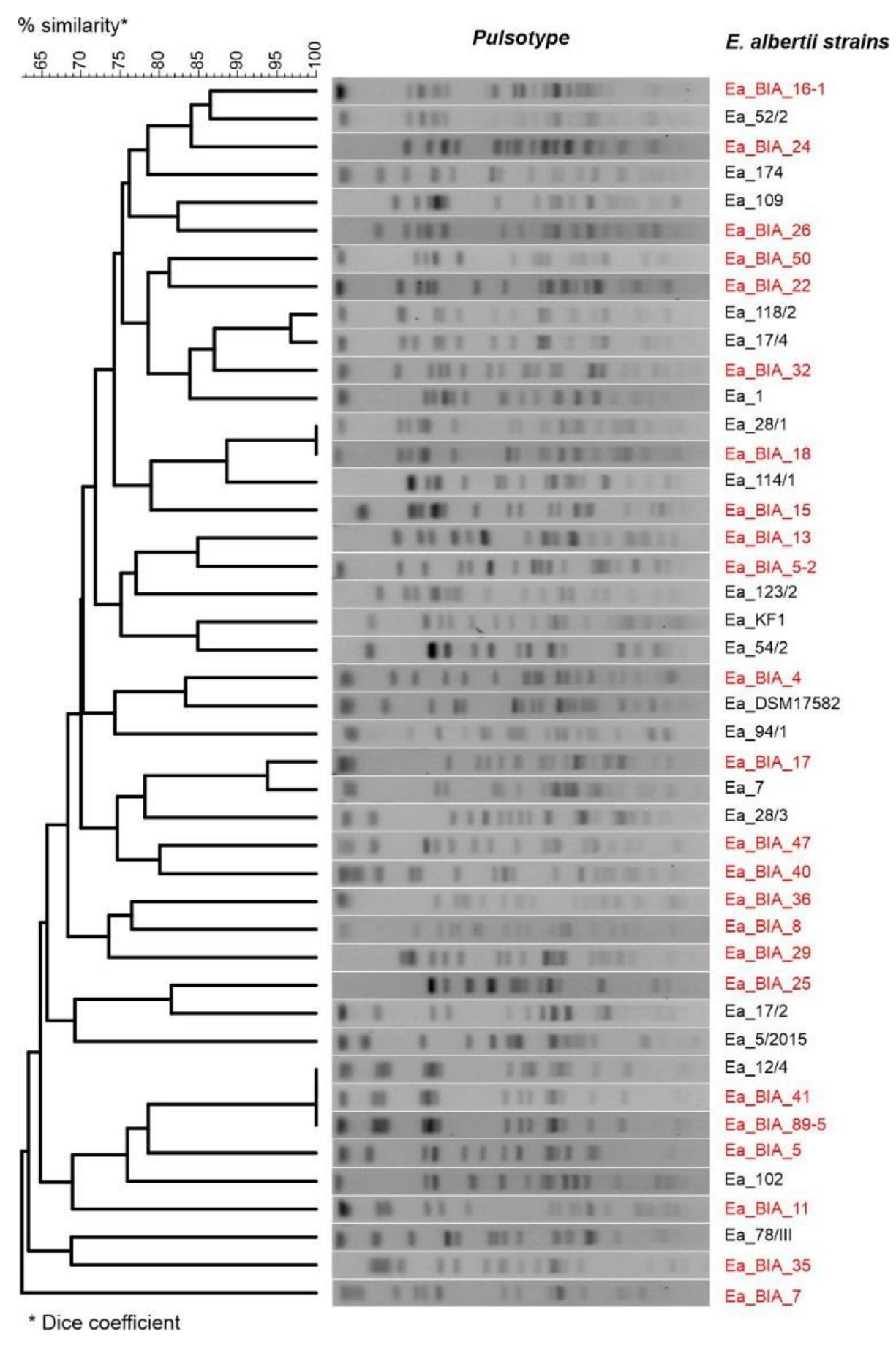
2.6. Restriction Fragment Length Polymorphism (RFLP)
2.7. Plasmid Profiles
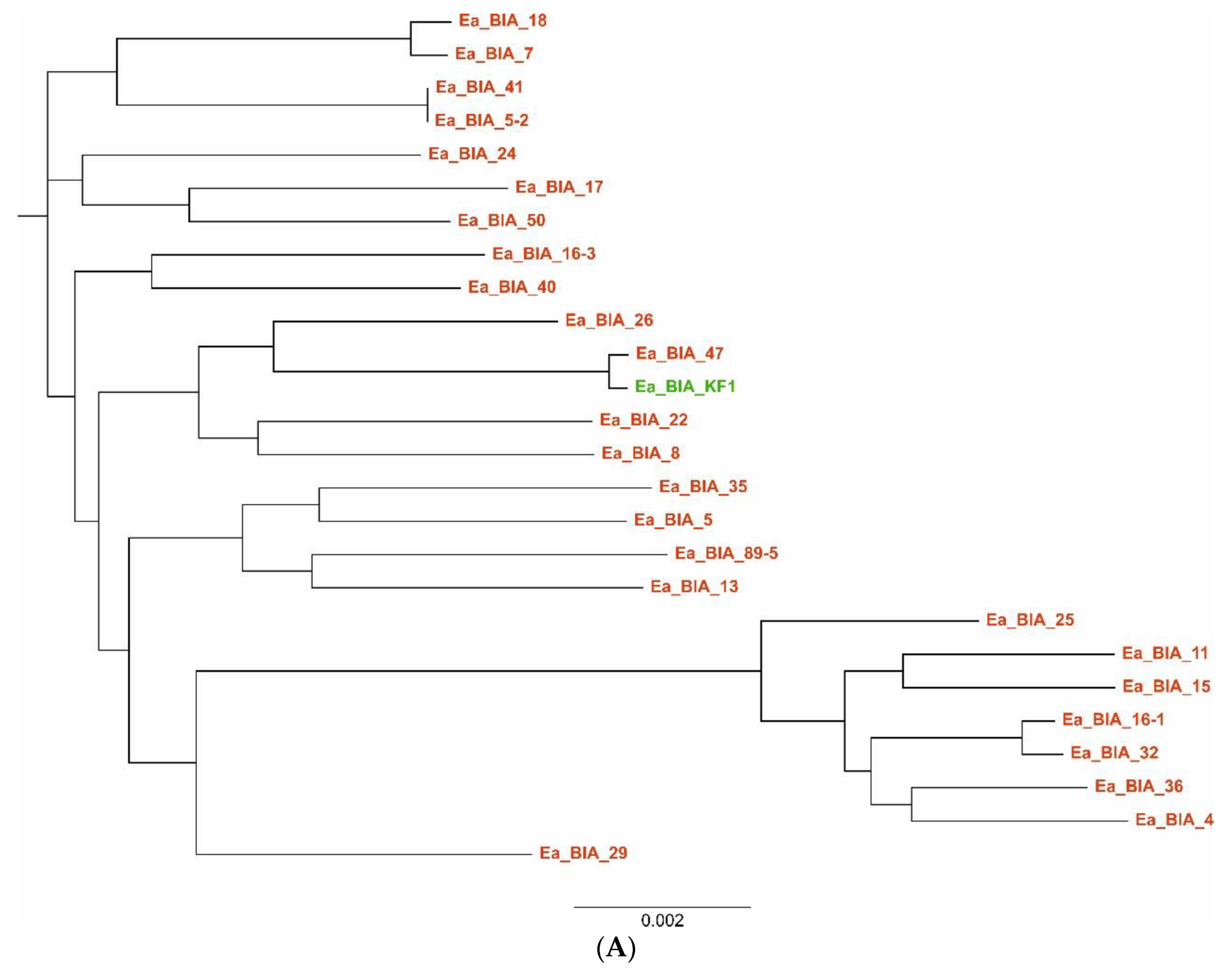
2.8. Multilocus Sequence Typing (MLST)
2.9. Virulence Gene Sequencing
2.10. Whole Genome Sequencing of E. Albertii Using Next-Generation Sequencing (NGS)
3. Data Analysis
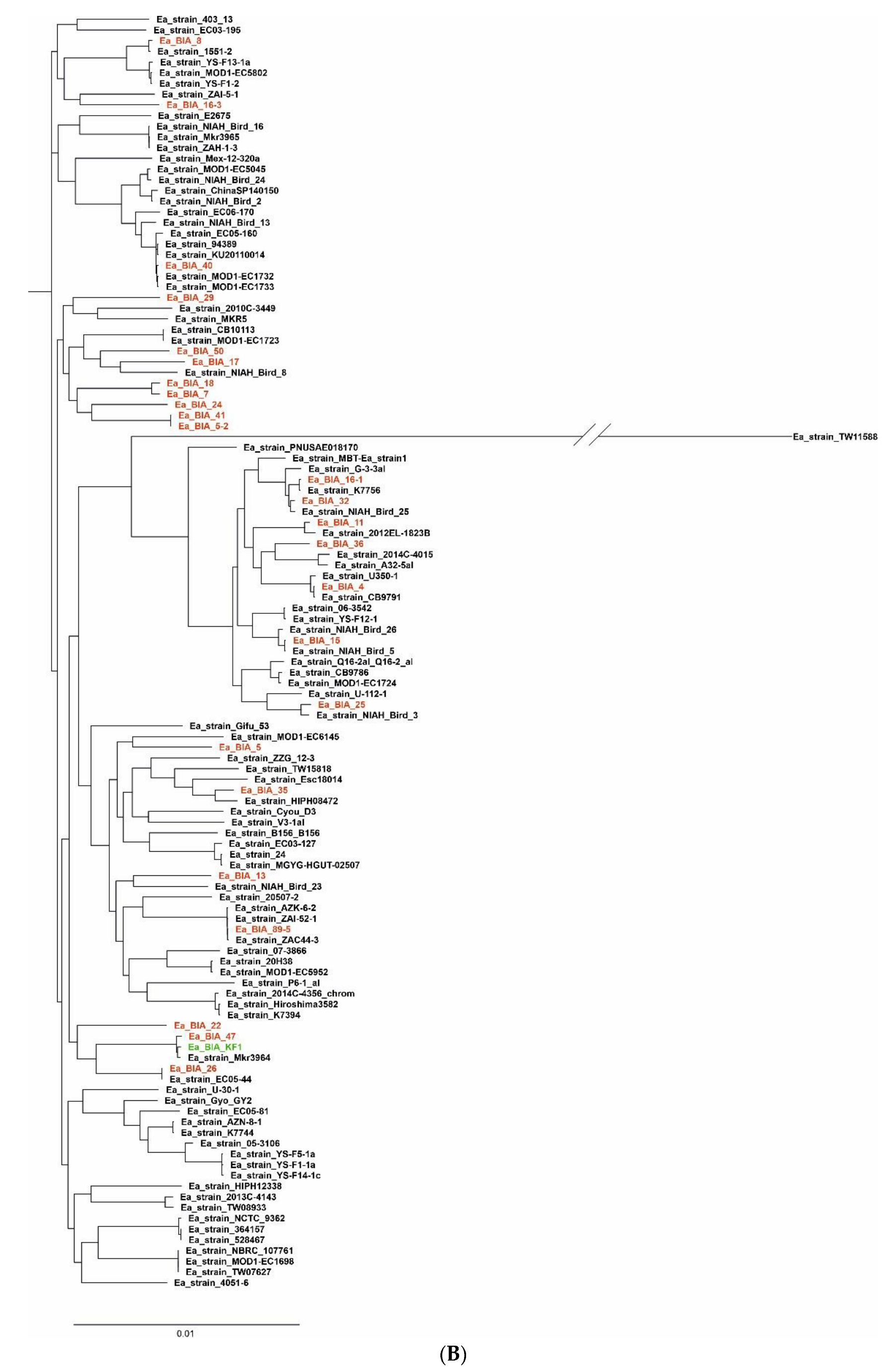
3.1. Bioinformatic Analysis and Study of E. albertii Genomes
3.1.1. Genome Annotation
3.1.2. Pangenome Estimation
3.2. Statistical Analysis
4. Results
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gordon, D.M. Reservoirs of Infection: The Epidemiological Characteristics of an Emerging Pathogen, Escherichia albertii; Research School of Biology. Australian National University: Canberra, Australia. Proceedings 2011, 61–73. Available online: https://www.aavac.com.au/files/2011-08.pdf (accessed on 27 April 2023).
- Nimri, L.F. Escherichia albertii, a newly emerging enteric pathogen with poorly defined properties. Diagn. Microbiol. Infect. Dis. 2013, 77, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Donnenberg, M.S.; Tzipori, S.; McKee, M.L.; O’Brien, A.D.; Alroy, I.J.; Kapert, J.B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Investig. 1993, 92, 1418–1424. [Google Scholar] [CrossRef] [PubMed]
- Asoshima, N.; Matsuda, M.; Shigemura, K.; Honda, M.; Yoshida, H.; Hiwaki, H.; Ogata, K.T. Identification of Escherichia albertii as a Causative Agent of a Food-Borne Outbreak Occurred in 2003. JPN J. Infect. Dis. 2014, 67, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Albert, M.J.; Alam, K.; Islam, M.; Montanaro, J.; Rahaman, A.S.; Haider, K.; Hossain, M.A.; Kibriya, A.K.; Tzipori, S. Hafnia alvei, a probable cause of diarrhea in humans. Infect. Immun. 1991, 59, 1507–1513. [Google Scholar] [CrossRef]
- Huys, G.; Cnockaert, M.; Janda, J.M.; Swings, J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 2003, 53, 807–810. [Google Scholar] [CrossRef]
- Oaks, J.L.; Besser, T.E.; Walk, S.T.; Gordon, D.M.; Beckmen, K.B.; Burek, K.A.; Haldorson, G.J.; Bradway, D.S.; Ouellette, L. Escherichia albertii in wild and domestic birds. Emerg. Infect. Dis. 2010, 16, 638–646. [Google Scholar] [CrossRef]
- Blanco, M.; Schumacher, S.; Tasara, T.; Zweifel, C.; Blanco, J.E.; Dahbi, G.; Blanco, J.; Stephan, R. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-eta2). BMC Microbiol. 2005, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Okuda, J.; Kurazono, H.; Takeda, Y. Distribution of the cytolethal distending toxin A gene (cdtA) among species of Shigella and Vibrio, and cloning and sequencing of the cdt gene from Shigella dysenteriae. Microb. Pathog. 1995, 18, 167–172. [Google Scholar] [CrossRef]
- Jinadasa, R.N.; Bloom, S.E.; Weiss, R.S.; Duhamel, G.E. Cytolethal distending toxin: A conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 2011, 157, 1851–1875. [Google Scholar] [CrossRef]
- Grasso, F.; Frisan, T. 2015. Bacterial genotoxins: Merging the DNA damage response into infection biology. Biomolecules 2015, 5, 1762–1782. [Google Scholar] [CrossRef]
- Taieb, F.; Petit, C.; Nougayrède, J.P.; Oswald, E. The enterobacterial genotoxins: Cytolethal distending toxin and colibactin. Ecosal. Plus 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Fedor, Y.; Vignard, J.; Nicolau-Travers, M.L.; Boutet-Robinet, E.; Watrin, C.; Salles, B.; Mirey, G. From single-strand breaks to double-strand breaks during S-phase: A new mode of action of the Escherichia coli cytolethal distending toxin. Cell Microbiol. 2013, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.D.; LaVeck, G.D. 1983. Purification and characterization of a Shigella dysenteriae 1-like toxin produced by Escherichia coli. Infect. Immun. 1983, 40, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Piérard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef]
- Ooka, T.; Seto, K.; Kawano, K.; Kobayashi, H.; Etoh, Y.; Ichihara, S.; Kaneko, A.; Isobe, J.; Yamaguchi, K.; Horikawa, K.; et al. Clinical significance of Escherichia albertii. Emerg Infect. Dis. 2012, 18, 488–492. [Google Scholar] [CrossRef]
- Murakami, K.; Etoh, Y.; Tanaka, E.; Ichihara, S.; Horikawa, K.; Kawano, K.; Ooka, T.; Kawamura, Y.; Ito, K. Shiga toxin 2f-producing Escherichia albertii from a symptomatic human. JPN J. Infect. Dis. 2014, 67, 204–20857. [Google Scholar] [CrossRef]
- Hinenoya, A.; Yasuda, N.; Hibino, T.; Shima, A.; Nagita, A.; Tsukamoto, T.; Shinji Yamasaki, S. Isolation and characterization of an Escherichia albertii strain producing three different toxins from a child with diarrhea. JPN J. Infect. Dis. 2017, 70, 252–257. [Google Scholar] [CrossRef]
- Brandal, L.T.; Tunsjø, H.S.; Ranheim, T.E.; Løbersli, I.; Lange, H.; Astrid Louise Wester, A.L. Shiga toxin 2a in Escherichia albertii. J. Clin. Microbiol. 2015, 53, 1454–1455. [Google Scholar] [CrossRef]
- Lynne, A.M.; Kariyawasam, S.; Wannemuehler, Y.; Johnson, T.J.; Johnson, S.J.; Sinha, A.S.; Lynne, D.K.; Moon, H.W.; Jordan, D.M.; Logue, C.M.; et al. Recombinant Iss as a potential vaccine for avian colibacillosis. Avian Dis. 2012, 56, 192–199. [Google Scholar] [CrossRef]
- Lima, M.P.; Yamamoto, D.; Santos, A.C.D.M.; Ooka, T.; Hernandes, R.T.; Vieira, M.A.; Gomes, T.A. Phenotypic characterization and virulence-related properties of Escherichia albertii strains isolated from children with diarrhea in Brazil. Pathog. Dis. 2019, 77, ftz014. [Google Scholar] [CrossRef]
- Fedoruk, K.; Daniluk, T.; Święcicka, I.; Murawska, E.; Sciepuk, M.; Leszczynska, K. First complete genome sequence of Escherichia albertii strain KF1, a new potential human enteric pathogen. Genom. Announc. 2014, 2, e00004–e00014. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutzb, F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Janben, T.; Schwarz, C.; Preikschat, P.; Voss, M.; Philipp, H.C.; Wieler, L.H. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. Int. J. Med. Microbiol. 2001, 291, 371–378. [Google Scholar] [CrossRef]
- Kahlmeter, G.; Brown, D.F.; Goldstein, F.W.; MacGowan, A.P.; Mouton, J.W.; Odenholt, I.; Rodloff, A.; Soussy, C.J.; Steinbakk, M.; Soriano, F.; et al. European Committee on Antimicrobial Susceptibility Testing (EUCAST) Technical Notes on antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2006, 12, 501–503. [Google Scholar] [CrossRef]
- Yang, J.L.; Wang, J.T.; Lauderdale, T.L.; Chang, S.C. Prevalence of extended-spectrum β-lactamases in Enterobacter cloacae in Taiwan and comparison of 3 phenotypic confirmatory methods for detecting extended-spectrum β-lactamase production. J. Microbiol. Immunol. Infect. 2009, 42, 310–316. [Google Scholar]
- Barton, B.M.; Harding, G.P.; Zuccarelli, A.J. A general method for detecting and sizing large plasmids. Anal. Biochem. 1995, 226, 235–240. [Google Scholar] [CrossRef]
- Stirling, D. DNA Extraction from Fungi, Yeast, and Bacteria. In PCR Protocols; Bartlett, J.M.S., Stirling, D., Eds.; Humana Press: Totowa, NJ, USA, 2003; pp. 53–54. [Google Scholar]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genom. Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Bryce, J.; Boschi-Pinto, C.; Shibuya, K.; Black, R.E. WHO Child Health Epidemiology Reference Group, 2005. WHO estimates of the causes of death in children. Lancet 2005, 365, 1147–1152. [Google Scholar] [CrossRef]
- Davidson, G.; Barnes, G.; Bass, D.; Cohen, M.; Fasano, A.; Fontaine, O.; Guandalini, S. Infectious diarrhea in children: Working Group Report of the First World Congress of Pediatric Gastroenterology, Hepatology, and Nutrition. J. Pediatr. Gastroenterol. Nutr. 2002, 35 (Suppl. 2), S143–S150. [Google Scholar] [CrossRef] [PubMed]
- Braden, C.R.; Tauxe, R.V. Emerging trends in foodborne diseases. Infect. Dis. Clin. North. Am. 2013, 27, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Inglis, T.J.; Merritt, A.J.; Bzdyl, N.; Urosevic, M.N. First bacteraemic human infection with Escherichia albertii. New Microbes New Infect. 2015, 8, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Ooka, T.; Tokuoka, E.; Furukawa, M. Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerg. Infect. Dis. 2013, 19, 144–146. [Google Scholar] [CrossRef]
- Masuda, K.; Ooka, T.; Akita, H.; Hiratsuka, T.; Takao, S.; Fukada, M.; Inoue, K.; Honda, M.; Toda, J.; Sugitani, W.; et al. Epidemiological Aspects of Escherichia albertii Outbreaks in Japan and Genetic Characteristics of the Causative Pathogen. Foodborne Pathog. Dis. 2020, 17, 144–150. [Google Scholar] [CrossRef]
- Afshin, N.; Bahareh, R.; Azadeh, F. Detection of Escherichia albertii in urinary and gastrointestinal infections in Kermanshah, Iran Research square. Int. J. Enteric Pathog. 2020, 9, 37–42. [Google Scholar]
- Zaki, M.E.S.; Eid, A.E.; El-Kazzaz, S.S.; El-Sabbagh, A.M. Molecular Study of Escherichia albertii in Pediatric Urinary Tract Infections. Open Microbiol. J. 2021, 15, 139–144. [Google Scholar] [CrossRef]
- Gomes, T.A.T.; Ooka, T.; Hernandes, R.T.; Yamamoto, D.; Hayashi, T. Escherichia albertii Pathogenesis. EcoSal. Plus 2020, 9, 1–18. [Google Scholar] [CrossRef]
- Stock, I.; Rahman, M.; Sherwood, K.J.; Wiedemann, B. Natural antimicrobial susceptibility patterns and biochemical identification of Escherichia albertii and Hafnia alvei strains. Diagn. Microbiol. Infect. Dis. 2005, 51, 151–163. [Google Scholar] [CrossRef]
- Hinenoya, A.; Li, X.; Zeng, X.; Sahin, O.; Moxley, R.A.; Logue, C.M.; Gillespie, B.; Yamasaki, S.; Lin, J. Isolation and characterization of Escherichia albertii in poultry at the pre-harvest level. Zoonoses Public Health 2021, 68, 213–225. [Google Scholar] [CrossRef]
- Konno, T.; Yatsuyangi, J.; Takahashi, S.; Kumagai, Y.; Wada, E.; Chiba, M.; Saito, S. Isolation and identification of Escherichia albertii from a patient in an outbreak of gastroenteritidis. JPN J. Infect. Dis. 2012, 65, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, H.; Xu, Y.; Bai, X.; Wang, J.; Zhang, Z.; Liu, X.; Miao, Y.; Zhang, L.; Li, X.; et al. Multidrug-resistant Escherichia albertii: Co-occurrence of β-lactamase and MCR-1 encoding genes. Front. Microbiol. 2018, 9, 258. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Wang, H.; Payne, M.J.; Liang, C.; Bai, L.; Zheng, H.; Zhang, Z.; Zhang, L.; Zhang, X.; Yan, G.; et al. Comparative genomics of Chinese and international isolates of Escherichia albertii: Population structure and evolution of virulence and antimicrobial resistance. Microb. Genom. 2021, 7, 000710. [Google Scholar] [CrossRef]
- Pennycott, T.W.; Ross, H.M.; McLaren, I.M.; Park, A.; Hopkins, G.F.; Foster, G. Causes of death of wild birds of the family Fringillidae in Britain. Vet. Rec. 1998, 143, 155–158. [Google Scholar] [CrossRef]
- Foster, G.; Ross, H.M.; Pennycott, T.W.; Hopkins, G.F.; McLaren, I.M. Isolation of Escherichia coli O86, K61 producing cyto-lethal distending toxin from wild birds of the finch family. Lett. Appl. Microbiol. 1998, 26, 395–398. [Google Scholar] [CrossRef]
- La Ragione, R.M.; McLaren, I.M.; Foster, G.; Cooley, W.A.; Woodward, M.J. Phenotypic and genotypic characterization of avian Escherichia coli O86, K61 isolates possessing a γ-like intimin. Appl. Environ. Microbiol. 2002, 68, 4932–4942. [Google Scholar] [CrossRef]
- Gordon, D.M.; Cowling, A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: Host and geographic effects. Microbiology 2003, 149, 3575–3586. [Google Scholar] [CrossRef]
- Maeda, E.; Murakami, K.; Sera, N.; Ito, K.; Fujimoto, S. Detection of Escherichia albertii from chicken meat and giblets. J. Vet. Med. Sci. 2015, 77, 871–873. [Google Scholar] [CrossRef]
- Barmettler, K.; Biggel, M.; Treier, A.; Muchaamba, F.; Vogler, B.R.; Stephan, R. Occurrence and Characteristics of Escherichia albertii in Wild Birds and Poultry Flocks in Switzerland. Microorganisms 2022, 10, 2265. [Google Scholar] [CrossRef] [PubMed]
- Asoshima, N.; Matsuda, M.; Shigemura, K.; Honda, M.; Yoshida, H.; Oda, T.; Hiwaki, H. Isolation of Escherichia albertii from raw chicken liver in Fukuoka City, Japan. Jpn J. Infect. Dis. 2015, 68, 248–250. [Google Scholar] [CrossRef]
- Wang, H.; Li, Q.; Bai, X.; Xu, Y.; Zhao, A.; Sun, H.; Deng, J.; Xiao, B.; Liu, X.; Sun, S.; et al. Prevalence of eae-positive, lactose non-fermenting Escherichia albertii from retail raw meat in China. Epidemiol. Infect. 2016, 144, 45–52. [Google Scholar] [CrossRef] [PubMed]


| E. albertii Strains | Antibiotics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AM | AMC | CXM | CEF | IP | CH | CIP | LEV | GN | SXT | |
| Ea_28/3; Ea_109; Ea_102; Ea_54/2; Ea_29/2; Ea_BIA_8; Ea_BIA_29; Ea_BIA_40; Ea_BIA_22; Ea_BIA_18; Ea_BIA_16-3; Ea_BIA_11; Ea_BIA_16-1; | R | S | I | S | S | S | S | S | S | S |
| Ea_174; Ea_28/1; Ea_78/III; Ea_7; Ea_15/2; Ea_17/4; Ea_5/2015; Ea_17/2; Ea_1; Ea_69/2; Ea_BIA_29; Ea_BIA_36; Ea_BIA_47; Ea_BIA_5; Ea_BIA_50; Ea_BIA_5-2; Ea_BIA_7; Ea_BIA_13; Ea_BIA_15; Ea_BIA_17; Ea_BIA_24; | S | S | I | S | S | S | S | S | S | S |
| Ea_12/4; Ea_94/2; Ea_BIA_89-5; Ea_BIA_26; | R | R | I | S | S | S | S | S | S | S |
| Ea_BIA_24 | R | S | I | S | S | R | S | S | S | S |
| Ea_BIA_4 | R | S | R | S | S | S | S | S | S | S |
| Ea_BIA_35 | S | S | R | S | S | S | R | S | S | S |
| Ea_BIA_40 | S | S | I | S | S | S | R | S | S | S |
| E. albertii Strains | Origin | Serotype | Type/Sequential Profile a | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ST | adk | fumC | gyrB | icd | mdh | purA | recA | |||
| Ea_BIA_11 | Bird (Sturnus vulgaris) | H33:O119 | 11608 | 228 | 293 | 85 | 207 | 116 | 119 | 106 |
| Ea_BIA_13 | Bird (Grus grus) | H52:O28 | nearest ST: 6059 | 163 | 395 | 134 | 386 | 378 | 444 | ~373 b |
| Ea_BIA_15 | Bird (Passer domesticus) | H52:O128 | 4736 | 228 | 170 | 85 | 332 | 116 | 149 | 106 |
| Ea_BIA_16-1 | Bird (Sturnus vulgaris) | H52:O128 | 5967 | 132 | 170 | 85 | 207 | 116 | 206 | 106 |
| Ea_BIA_16-3 | Bird (Sturnus vulgaris) | H5:- | nearest ST: 11622 | 228 | 788 | 85 | ~468 b | 378 | 149 | ~103 b |
| Ea_BIA_17 | Bird (Grus grus) | H52:- | nearest ST: 10710 | 133 | ~399 b | 85 | 189 | 142 | 141 | 176 |
| Ea_BIA_18 | Bird (Grus grus) | H52:O115 | nearest ST: 1846 | 133 | 298 | 235 | 255 | ~190 b | 66 | 176 |
| Ea_BIA_22 | Bird (Grus grus) | H52:- | nearest ST: 2683 c | 567 | ~394 b | 134 | 1787 | 255 | 113 | 226 |
| Ea_BIA_24 | Bird (Grus grus) | H52:O69 | nearest ST: 6059 c | 228 | ~788 b | ~85 b | 330 | 109 | ~69 b | 176 |
| Ea_BIA_25 | Bird (Grus grus) | H52:O1 | nearest ST: 1338 | ~132 b | 170 | 85 | 207 | 154 | 149 | 135 |
| Ea_BIA_26 | Bird (Grus grus) | H33:O182 | 6058 | 539 | 788 | 134 | 483 | 255 | 149 | 75 |
| Ea_BIA_29 | Bird (unidentified species) | H5:- | nearest ST: 11971 | 132 | 1587 | 134 | ~872 b | 256 | 66 | 76 |
| Ea_BIA_32 | Bird (Delichon urbicum) | H5:O8 | nearest ST: 5399 c | ~379 b | ~170 b | 85 | 207 | 116 | 206 | 106 |
| Ea_BIA_35 | Bird (Anser spp.) | H52:O152 | 10709 | 163 | 1283 | 852 | 189 | 68 | 66 | 225 |
| Ea_BIA_36 | Bird (unidentified species) | H33:O115 | 11195 | 228 | 1645 | 85 | 332 | 116 | 119 | 106 |
| Ea_BIA_4 | Bird (unidentified species) | H33:O1 | 6049 | 537 | 170 | 85 | 332 | 116 | 206 | 106 |
| Ea_BIA_40 | Bird (Anser spp.) | H52:- | 3762 | 132 | 394 | 134 | 144 | 110 | 66 | 102 |
| Ea_BIA_41 | Bird (Sturnus vulgaris) | H52:- | nearest ST: 12964 | 132 | 298 | 134 | ~189 b | 885 | 206 | ~226 b |
| Ea_BIA_47 | Backwaters (Jancewicze) | H33:- | 7365 | 695 | 111 | 85 | 483 | 109 | 149 | 75 |
| Ea_BIA_5 | Bird (unidentified species) | H52:- | 8691 | 163 | 393 | 85 | 189 | 68 | 246 | 125 |
| Ea_BIA_50 | Bird (Grus grus) | H52:O9 | nearest ST: 10710 c | ~393 b | 104 | ~85 b | ~545 b | 111 | 141 | ~176 b |
| Ea_BIA_5-2 | Bird (Columba livia) | H33:O9 | nearest ST: 12964 | 132 | 298 | 134 | ~189 b | 1238 | 206 | 885 |
| Ea_BIA_7 | Bird (Columba livia) | H52:O115 | 1846 | 163 | 298 | 235 | 255 | 190 | 66 | 176 |
| Ea_BIA_8 | Bird (Columba livia) | H52:O131 | 2681 | 134 | 164 | 85 | 169 | 254 | 69 | 103 |
| Ea_BIA_89-5 | Bird (Grus grus) | H52:O3 | 2680 | 293 | 395 | 85 | 330 | 68 | 248 | 226 |
| E. albertii Strains | Chromosome Size | Number of Genes | Number of Plasmids | Plasmid Size |
|---|---|---|---|---|
| Ea_BIA_11 | 4,779,775 | 4495 | 3 | 5952–110,945 |
| Ea_BIA_13 | 4,819,152 | 4503 | 7 | 1589–128,726 |
| Ea_BIA_15 | 4,772,054 | 4581 | 4 | 5140–130,064 |
| Ea_BIA_16-1 | 4,573,338 | 4248 | 3 | 2733–99,447 |
| Ea_BIA_16-3 | 4,838,644 | 4548 | 2 | 5055–82,983 |
| Ea_BIA_17 | 4,854,152 | 4728 | 3 | 1590–132,593 |
| Ea_BIA_18 | 4,871,752 | 4587 | 5 | 2760–144,030 |
| Ea_BIA_22 | 4,884,955 | 4718 | 8 | 1590–152,064 |
| Ea_BIA_24 | 4,841,726 | 4513 | 7 | 1589–136,544 |
| Ea_BIA_25 | 4,810,099 | 4480 | 4 | 1589–100,741 |
| Ea_BIA_26 | 4,654,873 | 4279 | 4 | 4703–114,713 |
| Ea_BIA_29 | 4,687,526 | 4362 | 2 | 4593–116,006 |
| Ea_BIA_32 | 4,822,417 | 4570 | 5 | 3758–125,136 |
| Ea_BIA_35 | 4,725,775 | 4422 | 4 | 2571–88,900 |
| Ea_BIA_36 | 4,790,901 | 4588 | 4 | 40,185–103,275 |
| Ea_BIA_4 | 4,776,253 | 4548 | 4 | 36,239–97,069 |
| Ea_BIA_40 | 4,826,552 | 4550 | 3 | 2719–95,160 |
| Ea_BIA_41 | 4,677,218 | 4327 | 2 | 13,675–76,945 |
| Ea_BIA_47 | 4,595,293 | 4288 | 2 | 2720–40,979 |
| Ea_BIA_5 | 4,782,897 | 4330 | 4 | 3909–143,188 |
| Ea_BIA_50 | 4,722,615 | 4365 | 4 | 1627–165,614 |
| Ea_BIA_5-2 | 4,681,166 | 4356 | 1 | 76,945 |
| Ea_BIA_7 | 4,960,105 | 4706 | 5 | 2283–177,310 |
| Ea_BIA_8 | 4,709,836 | 4505 | 3 | 47,425–108,071 |
| Ea_BIA_89-5 | 5,141,010 | 4990 | 1 | 75,610 |
| (A) | ||
|---|---|---|
| Type Genes | No. Strains (N = 26 *) | No. Genes |
| Core genes | (99% ≤ strains ≤ 100%) | 2966 |
| Soft core genes | (95% ≤ strains < 99%) | 252 |
| Shell genes | (15% ≤ strains < 95%) | 3062 |
| Cloud genes | (0% ≤ strains < 15%) | 7888 |
| Total | (0% ≤ strains ≤ 100%) | 14,168 |
| * included previous E. albertii isolate from our center was used (DOI: 10.1128/genomeA.00004-14). | ||
| (B) | ||
| Type Genes | No. Strains (N = 119) | No. Genes |
| Core genes | (99% ≤ strains ≤ 100%) | 2088 |
| Soft core genes | (95% ≤ strains < 99%) | 511 |
| Shell genes | (15% ≤ strains < 95%) | 3052 |
| Cloud genes | (0% ≤ strains < 15%) | 19,506 |
| Total | (0% ≤ strains ≤ 100%) | 25,157 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leszczyńska, K.; Święcicka, I.; Daniluk, T.; Lebensztejn, D.; Chmielewska-Deptuła, S.; Leszczyńska, D.; Gawor, J.; Kliber, M. Escherichia albertii as a Potential Enteropathogen in the Light of Epidemiological and Genomic Studies. Genes 2023, 14, 1384. https://doi.org/10.3390/genes14071384
Leszczyńska K, Święcicka I, Daniluk T, Lebensztejn D, Chmielewska-Deptuła S, Leszczyńska D, Gawor J, Kliber M. Escherichia albertii as a Potential Enteropathogen in the Light of Epidemiological and Genomic Studies. Genes. 2023; 14(7):1384. https://doi.org/10.3390/genes14071384
Chicago/Turabian StyleLeszczyńska, Katarzyna, Izabela Święcicka, Tamara Daniluk, Dariusz Lebensztejn, Sylwia Chmielewska-Deptuła, Dorota Leszczyńska, Jan Gawor, and Małgorzata Kliber. 2023. "Escherichia albertii as a Potential Enteropathogen in the Light of Epidemiological and Genomic Studies" Genes 14, no. 7: 1384. https://doi.org/10.3390/genes14071384
APA StyleLeszczyńska, K., Święcicka, I., Daniluk, T., Lebensztejn, D., Chmielewska-Deptuła, S., Leszczyńska, D., Gawor, J., & Kliber, M. (2023). Escherichia albertii as a Potential Enteropathogen in the Light of Epidemiological and Genomic Studies. Genes, 14(7), 1384. https://doi.org/10.3390/genes14071384






