Translation Arrest: A Key Player in Plant Antiviral Response
Abstract
1. Introduction
2. Translation of Viral Transcripts in Plants
2.1. Adaptations of Viral RNAs for Enhanced Translation
2.2. Viral Translation
2.3. Optimization of Viral Translation
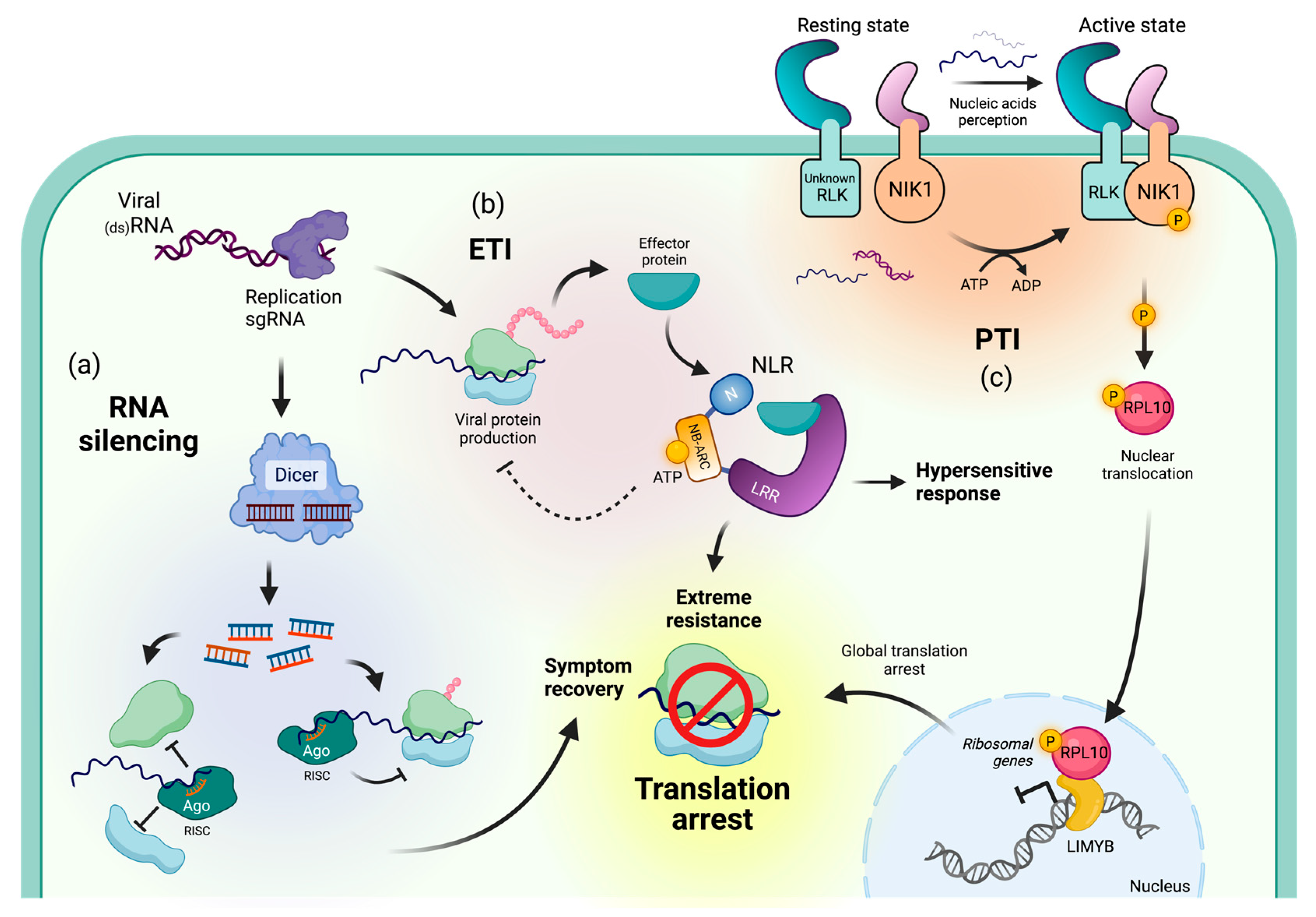
3. Immune Mechanisms Resulting in Translational Repression of Viral RNAs
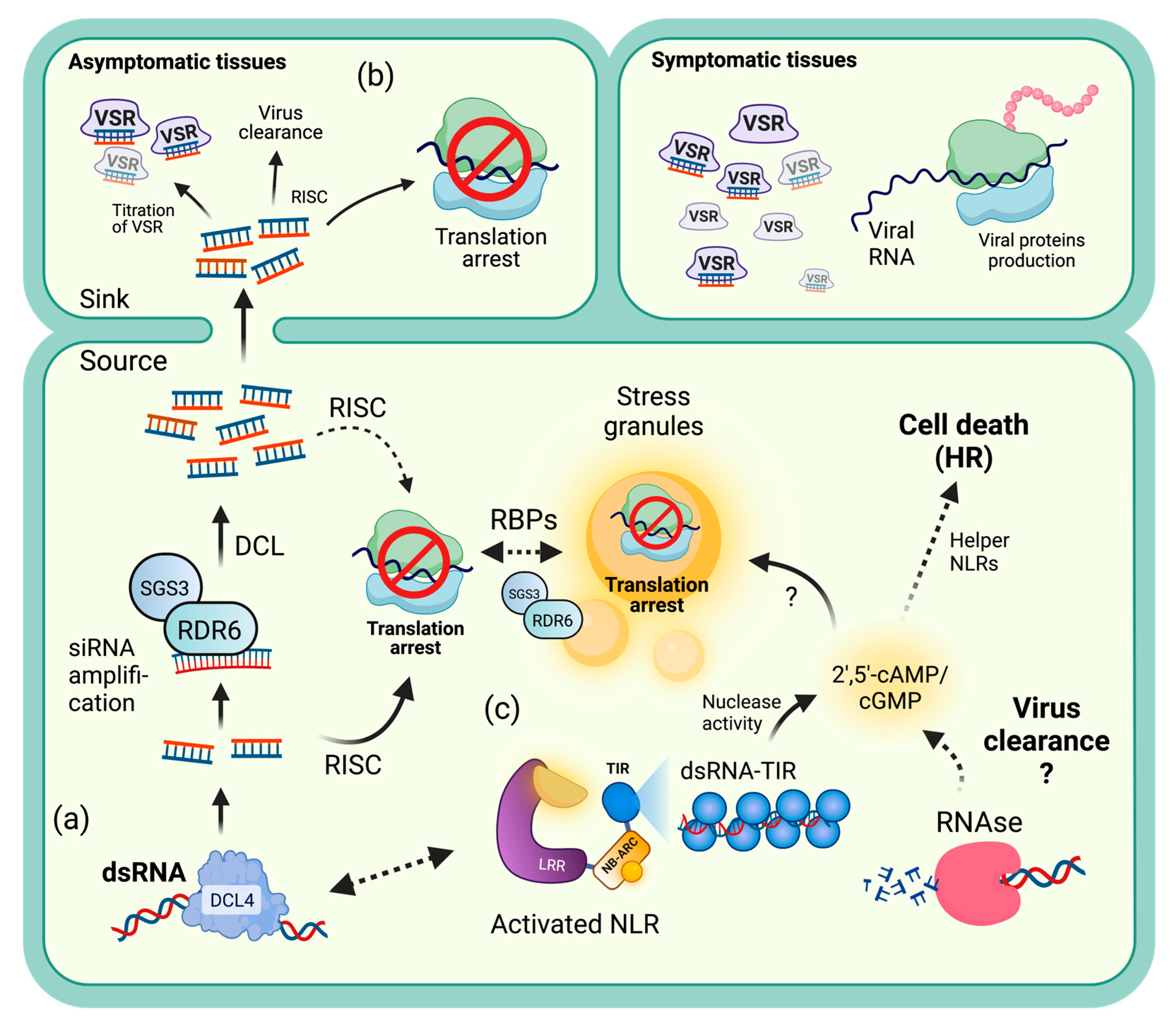
3.1. Viral Recovery through PTGS-Mediated Translational Repression
3.2. NLR-Mediated Translational Arrest
4. Translational Repression
4.1. Transcript Recognition
4.2. Translation Inhibition
4.3. Processing Bodies
4.4. Translation Repression Depends on AGOs and VSR Interference
5. Discussion
5.1. Similarities between TRV Recovery and N-Mediated Translational Repression
5.2. Processing Bodies
5.3. Gene Regulation during NLR-Mediated TA
5.4. Transcript Recognition during NLR-Mediated TA
5.5. Translation Inhibition Hypothesis
6. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging Infectious Diseases of Plants: Pathogen Pollution, Climate Change and Agrotechnology Drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.C.; Adamczyk, J.; Rinderer, T.; Yao, J.; Danka, R.; Luttrell, R.; Gore, J. Spray Toxicity and Risk Potential of 42 Commonly Used Formulations of Row Crop Pesticides to Adult Honey Bees (Hymenoptera: Apidae). J. Econ. Entomol. 2015, 108, 2640–2647. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.S.S. Plant Resistance to Viruses. In Encyclopedia of Virology; Granoff, A., Webster, R.G., Eds.; Academic Press: San Diego, CA, USA, 2021; pp. 1300–1307. [Google Scholar]
- Albar, L.; Bangratz-Reyser, M.; Hébrard, E.; Ndjiondjop, M.N.; Jones, M.; Ghesquière, A. Mutations in the EIF(Iso)4G Translation Initiation Factor Confer High Resistance of Rice to Rice Yellow Mottle Virus. Plant J. 2006, 47, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Muhsin, M.; Atienza, G.A.; Kwak, D.Y.; Kim, S.M.; Leon, T.B.D.; Angeles, E.R.; Coloquio, E.; Kondoh, H.; Satoh, K.; et al. Single Nucleotide Polymorphisms in a Gene for Translation Initiation Factor (EIF4G) of Rice (Oryza sativa) Associated with Resistance to Rice Tungro Spherical Virus. Mol. Plant-Microbe Interact. 2009, 23, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Takakura, Y.; Udagawa, H.; Shinjo, A.; Koga, K. Mutation of a Nicotiana tabacum L. Eukaryotic Translation-Initiation Factor Gene Reduces Susceptibility to a Resistance-Breaking Strain of Potato Virus Y. Mol. Plant Pathol. 2018, 19, 2124–2133. [Google Scholar] [CrossRef]
- Van Schie, C.C.N.; Takken, F.L.W. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Kryovrysanaki, N.; James, A.; Tselika, M.; Bardani, E.; Kalantidis, K. RNA silencing pathway in plant development and defense. Int. J. Dev. Biol. 2022, 66, 163–175. [Google Scholar] [CrossRef]
- Derrien, B.; Baumberger, N.; Schepetilnikov, M.; Viotti, C.; De Cillia, J.; Ziegler-Graff, V.; Isono, E.; Schumacher, K.; Genschik, P. Degradation of the Antiviral Component ARGONAUTE1 by the Autophagy Pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 15942–15946. [Google Scholar] [CrossRef]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with Host SGS3 Is Required for Suppression of RNA Silencing by Tomato Yellow Leaf Curl Virus V2 Protein. Proc. Natl. Acad. Sci. USA 2008, 105, 157–161. [Google Scholar] [CrossRef]
- Guo, H.; Song, X.; Xie, C.; Huo, Y.; Zhang, F.; Chen, X.; Geng, Y.; Fang, R. Rice Yellow Stunt Rhabdovirus Protein 6 Suppresses Systemic RNA Silencing by Blocking RDR6-Mediated Secondary SiRNA Synthesis. Mol. Plant-Microbe Interact. 2013, 26, 927–936. [Google Scholar] [CrossRef]
- Haas, G.; Azevedo, J.; Moissiard, G.; Geldreich, A.; Himber, C.; Bureau, M.; Fukuhara, T.; Keller, M.; Voinnet, O. Nuclear Import of CaMV P6 Is Required for Infection and Suppression of the RNA Silencing Factor DRB4. EMBO J. 2008, 27, 2102–2112. [Google Scholar] [CrossRef]
- Schott, G.; Mari-Ordonez, A.; Himber, C.; Alioua, A.; Voinnet, O.; Dunoyer, P. Differential Effects of Viral Silencing Suppressors on SiRNA and MiRNA Loading Support the Existence of Two Distinct Cellular Pools of ARGONAUTE1. EMBO J. 2012, 31, 2553–2565. [Google Scholar] [CrossRef] [PubMed]
- Várallyay, É.; Válóczi, A.; Ágyi, Á.; Burgyán, J.; Havelda, Z. Plant Virus-Mediated Induction of MiR168 Is Associated with Repression of ARGONAUTE1 Accumulation. EMBO J. 2010, 29, 3507–3519. [Google Scholar] [CrossRef] [PubMed]
- Vogler, H.; Akbergenov, R.; Shivaprasad, P.V.; Dang, V.; Fasler, M.; Kwon, M.-O.; Zhanybekova, S.; Hohn, T.; Heinlein, M. Modification of Small RNAs Associated with Suppression of RNA Silencing by Tobamovirus Replicase Protein. J. Virol. 2007, 81, 10379–10388. [Google Scholar] [CrossRef]
- Sett, S.; Prasad, A.; Prasad, M. Resistance Genes on the Verge of Plant–Virus Interaction. Trends Plant Sci. 2022, 27, 1242–1252. [Google Scholar] [CrossRef]
- Collier, S.M.; Moffett, P. NB-LRRs Work a “Bait and Switch” on Pathogens. Trends Plant Sci. 2009, 14, 521–529. [Google Scholar] [CrossRef]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2017, 219, 17–24. [Google Scholar] [CrossRef]
- Takken, F.L.; Albrecht, M.; Tameling, W.I. Resistance Proteins: Molecular Switches of Plant Defence. Curr. Opin. Plant Biol. 2006, 9, 383–390. [Google Scholar] [CrossRef]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx Gene from Potato Controls Separate Virus Resistance and Cell Death Responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Ghazala, W.; Varrelmann, M. Tobacco Rattle Virus 29K Movement Protein Is the Elicitor of Extreme and Hypersensitive-like Resistance in Two Cultivars of Solanum tuberosum. Mol. Plant-Microbe Interact. 2007, 20, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Grech-Baran, M.; Witek, K.; Szajko, K.; Witek, A.I.; Morgiewicz, K.; Wasilewicz-Flis, I.; Jakuczun, H.; Marczewski, W.; Jones, J.D.G.; Hennig, J. Extreme Resistance to Potato Virus Y in Potato Carrying the Rysto Gene Is Mediated by a TIR-NLR Immune Receptor. Plant Biotechnol. J. 2020, 18, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Sukarta, O.C.A.; Townsend, P.D.; Llewelyn, A.; Dixon, C.H.; Slootweg, E.J.; Pålsson, L.O.; Takken, F.L.W.; Goverse, A.; Cann, M.J. A DNA-Binding Bromodomain-Containing Protein Interacts with and Reduces Rx1-Mediated Immune Response to Potato Virus X. Plant Commun. 2020, 1, 100086. [Google Scholar] [CrossRef]
- Alazem, M.; Tseng, K.C.; Chang, W.C.; Seo, J.K.; Kim, K.H. Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses 2018, 10, 581. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Zamora, A.; Azhar, M.T.; Sacco, M.A.; Lambert, L.H.; Moffett, P. Virus Resistance Induced by NB–LRR Proteins Involves Argonaute4-Dependent Translational Control. Plant J. 2009, 58, 940–951. [Google Scholar] [CrossRef]
- Chowdhury, R.N.; Lasky, D.; Karki, H.; Zhang, Z.; Goyer, A.; Halterman, D.; Rakotondrafara, A.M. HCPRO Suppression of Callose Deposition Contributes to Strain-Specific Resistance against Potato Virus Y. Phytopathology 2020, 110, 164–173. [Google Scholar] [CrossRef]
- Iglesias, V.A.; Meins, F. Movement of Plant Viruses Is Delayed in a β-1,3-Glucanase-Deficient Mutant Showing a Reduced Plasmodesmatal Size Exclusion Limit and Enhanced Callose Deposition. Plant J. 2000, 21, 157–166. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Knip, M.; Schachtschabel, J.; Beijaert, M.S.; Takken, F.L.W. Perturbation of Nuclear–Cytosolic Shuttling of Rx1 Compromises Extreme Resistance and Translational Arrest of Potato Virus X Transcripts. Plant J. 2021, 106, 468–479. [Google Scholar] [CrossRef]
- Ross, B.T.; Zidack, N.K.; Flenniken, M.L. Extreme Resistance to Viruses in Potato and Soybean. Front. Plant Sci. 2021, 12, 658981. [Google Scholar] [CrossRef]
- Seo, J.K.; Kwon, S.J.; Cho, W.K.; Choi, H.S.; Kim, K.H. Type 2C Protein Phosphatase Is a Key Regulator of Antiviral Extreme Resistance Limiting Virus Spread. Sci. Rep. 2014, 4, srep05905. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.E.; Jones, R.A.C.; Coutts, R.H.A. Expression of Potato Virus X Resistance Gene Rx in Potato Leaf Protoplasts. J. Gen. Virol. 1986, 67, 2341–2345. [Google Scholar] [CrossRef]
- Bendahmane, A.; Köhm, B.A.; Dedi, C.; Baulcombe, D.C. The Coat Protein of Potato Virus X Is a Strain-Specific Elicitor of Rx1-Mediated Virus Resistance in Potato. Plant J. 1995, 8, 933–941. [Google Scholar] [CrossRef]
- Mestre, P.; Baulcombe, D.C. Elicitor-Mediated Oligomerization of the Tobacco N Disease Resistance Protein. Plant Cell 2006, 18, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Szittya, G.; Molnár, A.; Silhavy, D.; Hornyik, C.; Burgyán, J. Short Defective Interfering RNAs of Tombusviruses Are Not Targeted but Trigger Post-Transcriptional Gene Silencing against Their Helper Virus. Plant Cell 2002, 14, 359–372. [Google Scholar] [CrossRef]
- Sotta, N.; Chiba, Y.; Miwa, K.; Takamatsu, S.; Tanaka, M.; Yamashita, Y.; Naito, S.; Fujiwara, T. Global Analysis of Boron-Induced Ribosome Stalling Reveals Its Effects on Translation Termination and Unique Regulation by AUG-Stops in Arabidopsis Shoots. Plant J. 2021, 106, 1455–1467. [Google Scholar] [CrossRef]
- Teixeira, D.; Sheth, U.; Valencia-Sanchez, M.A.; Brengues, M.; Parker, R. Processing Bodies Require RNA for Assembly and Contain Nontranslating MRNAs. RNA 2005, 11, 371–382. [Google Scholar] [CrossRef]
- Wu, H.; Li, B.; Iwakawa, H.O.; Pan, Y.; Tang, X.; Ling-hu, Q.; Liu, Y.; Sheng, S.; Feng, L.; Zhang, H.; et al. Plant 22-Nt SiRNAs Mediate Translational Repression and Stress Adaptation. Nature 2020, 581, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread Translational Inhibition by Plant MiRNAs and SiRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Meteignier, L.V.; Zhou, J.; Cohen, M.; Bhattacharjee, S.; Brosseau, C.; Chan, M.G.C.; Robatzek, S.; Moffett, P. NB-LRR Signaling Induces Translational Repression of Viral Transcripts and the Formation of RNA Processing Bodies through Mechanisms Differing from Those Activated by UV Stress and RNAi. J. Exp. Bot. 2016, 67, 2353–2366. [Google Scholar] [CrossRef]
- Teixeira, R.M.; Ferreira, M.A.; Raimundo, G.A.S.; Loriato, V.A.P.; Reis, P.A.B.; Fontes, E.P.B. Virus Perception at the Cell Surface: Revisiting the Roles of Receptor-like Kinases as Viral Pattern Recognition Receptors. Mol. Plant Pathol. 2019, 20, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Zorzatto, C.; MacHado, J.P.B.; Lopes, K.V.G.; Nascimento, K.J.T.; Pereira, W.A.; Brustolini, O.J.B.; Reis, P.A.B.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-Mediated Translation Suppression Functions as a Plant Antiviral Immunity Mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef]
- Ghoshal, B.; Sanfaçon, H. Symptom Recovery in Virus-Infected Plants: Revisiting the Role of RNA Silencing Mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Nicole, M.C.; Meteignier, L.V.; Hong, N.; Wang, G.; Moffett, P. Different Roles for RNA Silencing and RNA Processing Components in Virus Recovery and Virus-Induced Gene Silencing in Plants. J. Exp. Bot. 2015, 66, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.P.B.; Calil, I.P.; Santos, A.A.; Fontes, E.P.B. Translational Control in Plant Antiviral Immunity. Genet. Mol. Biol. 2017, 40, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Passmore, L.A.; Coller, J. Roles of MRNA Poly(A) Tails in Regulation of Eukaryotic Gene Expression. Nat. Rev. Mol. Cell Biol. 2022, 23, 93–106. [Google Scholar] [CrossRef]
- Geng, G.; Wang, D.; Liu, Z.; Wang, Y.; Zhu, M.; Cao, X.; Yu, C.; Yuan, X. Translation of Plant RNA Viruses. Viruses 2021, 13, 2499. [Google Scholar] [CrossRef]
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Comparative Genetics: Systematic Discovery of Cap-Independent Translation Sequences in Human and Viral Genomes. Science 2016, 351, aad4939. [Google Scholar] [CrossRef]
- Jang, S.K.; Krausslich, H.-G.; Nicklin, M.J.H.; Duke, G.M.; Palmenberg, A.C.; Wimmer’, E. A Segment of the 5′ Nontranslated Region of Encephalomyocarditis Virus RNA Directs Internal Entry of Ribosomes during in Vitro Translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef]
- Karetnikov, A.; Lehto, K. The RNA2 5′ Leader of Blackcurrant Reversion Virus Mediates Effecient in Vivo Translation through an Internal Ribosomal Entry Site Mechanism. J. Gen. Virol. 2007, 88, 286–297. [Google Scholar] [CrossRef]
- Roberts, R.; Mayberry, L.K.; Browning, K.S.; Rakotondrafara, A.M. The Triticum Mosaic Virus 5′ Leader Binds to Both EIF4G and EIFiso4G for Translation. PLoS ONE 2017, 12, e0169602. [Google Scholar] [CrossRef]
- Khan, M.A.; Miyoshi, H.; Gallie, D.R.; Goss, D.J. Potyvirus Genome-Linked Protein, VPg, Directly Affects Wheat Germ in Vitro Translation: Interactions with Translation Initiation Factors EIF4F and EIFiso4F. J. Biol. Chem. 2008, 283, 1340–1349. [Google Scholar] [CrossRef]
- Miyoshi, H.; Okade, H.; Muto, S.; Suehiro, N.; Nakashima, H.; Tomoo, K.; Natsuaki, T. Turnip Mosaic Virus VPg Interacts with Arabidopsis thaliana EIF(Iso)4E and Inhibits in Vitro Translation. Biochimie 2008, 90, 1427–1434. [Google Scholar] [CrossRef]
- Duijsings, D.; Kormelink, R.; Goldbach, R. Alfalfa Mosaic Virus RNAs Serve as Cap Donors for Tomato Spotted Wilt Virus Transcription during Coinfection of Nicotiana benthamiana. J. Virol. 1999, 73, 5172–5175. [Google Scholar] [CrossRef]
- Estabrook, E.M.; Tsai, J.; Falk, B.W. In Vivo Transfer of Barley Stripe Mosaic Hordeivirus Ribonucleotides to the 5′ Terminus of Maize Stripe Tenuivirus RNAs. Proc. Natl. Acad. Sci. USA 1998, 95, 8304–8309. [Google Scholar] [CrossRef] [PubMed]
- Kneller, E.L.P.; Rakotondrafara, A.M.; Miller, W.A. Cap-Independent Translation of Plant Viral RNAs. Virus Res. 2006, 119, 63–75. [Google Scholar] [CrossRef]
- Miller, W.A.; Koev, G. Synthesis of Subgenomic RNAs by Positive-Strand RNA Viruses. Virology 2000, 273, 1–8. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Korniy, N.; Klimova, M.; Karki, P.; Peng, B.Z.; Senyushkina, T.; Belardinelli, R.; Maracci, C.; Wohlgemuth, I.; Samatova, E.; et al. Translational Recoding: Canonical Translation Mechanisms Reinterpreted. Nucleic Acids Res. 2020, 48, 1056–1067. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Dinman, J.D.; Green, R. Translation Elongation and Recoding in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032649. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Maule, A.J. Inhibition of Host Gene Expression Associated with Plant Virus Replication. Science 1995, 267, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Jan, E.; Mohr, I.; Walsh, D. A Cap-to-Tail Guide to MRNA Translation Strategies in Virus-Infected Cells. Annu. Rev. Virol. 2016, 3, 283–307. [Google Scholar] [CrossRef]
- Fernández de Castro, I.; Tenorio, R.; Risco, C. Virus Factories. In Encyclopedia of Virology; Bamford, D.A., Zuckerman, M., Eds.; Academic Press: San Diego, CA, USA, 2021; pp. 495–500. [Google Scholar] [CrossRef]
- Miras, M.; Truniger, V.; Querol-Audi, J.; Aranda, M.A. Analysis of the Interacting Partners EIF4F and 3′-CITE Required for Melon Necrotic Spot Virus Cap-Independent Translation. Mol. Plant Pathol. 2017, 18, 635–648. [Google Scholar] [CrossRef]
- Nicholson, B.L.; Wu, B.; Chevtchenko, I.; White, K.A. Tombusvirus Recruitment of Host Translational Machinery via the 3′ UTR. RNA 2010, 16, 1402–1419. [Google Scholar] [CrossRef]
- Royall, E.; Doyle, N.; Abdul-Wahab, A.; Emmott, E.; Morley, S.J.; Goodfellow, I.; Roberts, L.O.; Locker, N. Murine Norovirus 1 (MNV1) Replication Induces Translational Control of the Host by Regulating EIF4E Activity during Infection. J. Biol. Chem. 2015, 290, 4748–4758. [Google Scholar] [CrossRef]
- Khan, M.A. Phosphorylation of Translation Initiation Factor EIFiso4E Promotes Translation through Enhanced Binding to Potyvirus VPg. J. Biochem. 2019, 165, 167–176. [Google Scholar] [CrossRef]
- Khan, M.A.; Kumar, P.; Akif, M.; Miyoshi, H. Phosphorylation of Eukaryotic Initiation Factor EIFiso4E Enhances the Binding Rates to VPg of Turnip Mosaic Virus. PLoS ONE 2021, 16, e0259688. [Google Scholar] [CrossRef]
- Hatta, T.; Bullivant, S.; Matthews, R.E.F. Fine Structure of Vesicles Induced in Chloroplasts of Chinese Cabbage Leaves by Infection with Turnip Yellow Mosaic Virus. J. Gen. Virol. 1973, 20, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S. An Ultrastructural Study of Inclusions and Disease Development in Plant Cells Infected by Cowpea Chlorotic Mottle Virus. J. Gen. Virol. 1977, 35, 535–543. [Google Scholar] [CrossRef]
- McCartney, A.W.; Greenwood, J.S.; Fabian, M.R.; White, K.A.; Mullen, R.T. Localization of the Tomato Bushy Stunt Virus Replication Protein P33 Reveals a Peroxisome-to-Endoplasmic Reticulum Sorting Pathway. Plant Cell 2005, 17, 3513–3531. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Hirai, K.; Kanda, A.; Ohnishi, J.; Ohki, T.; Tsuda, S. Induction of Necrosis via Mitochondrial Targeting of Melon Necrotic Spot Virus Replication Protein P29 by Its Second Transmembrane Domain. Virology 2009, 390, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Schaad, M.C.; Jensen, P.E.; Carrington, J.C. Formation of Plant RNA Virus Replication Complexes on Membranes: Role of an Endoplasmic Reticulum-Targeted Viral Protein. EMBO J. 1997, 16, 4049–4059. [Google Scholar] [CrossRef]
- Amari, K.; Lerich, A.; Schmitt-Keichinger, C.; Dolja, V.V.; Ritzenthaler, C. Tubule-Guided Cell-to-Cell Movement of a Plant Virus Requires Class XI Myosin Motors. PLoS Pathog. 2011, 7, e1002327. [Google Scholar] [CrossRef]
- Katsafanas, G.C.; Moss, B. Colocalization of Transcription and Translation within Cytoplasmic Poxvirus Factories Coordinates Viral Expression and Subjugates Host Functions. Cell Host Microbe 2007, 2, 221–228. [Google Scholar] [CrossRef]
- Molnár, A.; Csorba, T.; Lakatos, L.; Várallyay, É.; Lacomme, C.; Burgyán, J. Plant Virus-Derived Small Interfering RNAs Originate Predominantly from Highly Structured Single-Stranded Viral RNAs. J. Virol. 2005, 79, 7812–7818. [Google Scholar] [CrossRef]
- Li, F.; Ding, S.-W. Virus Counterdefense: Diverse Strategies for Evading the RNA-Silencing Immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef]
- Ding, S.-W.; Lu, R. Virus-Derived SiRNAs and PiRNAs in Immunity and Pathogenesis. Curr. Opin. Virol. 2011, 1, 533–544. [Google Scholar] [CrossRef]
- Liu, Q.; Feng, Y.; Zhu, Z. Dicer-like (DCL) Proteins in Plants. Funct. Integr. Genomics 2009, 9, 277–286. [Google Scholar] [CrossRef]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical Action and Inhibition of Plant Dicer-like Proteins in Antiviral Defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Yang, M.; Le, B.H.; He, W.; Hou, Y. The Master Role of SiRNAs in Plant Immunity. Mol. Plant Pathol. 2022, 23, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral Roles of Plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, Y.-R.; Pei, Y.; Lin, S.-S.; Tuschl, T.; Patel, D.J.; Chua, N.-H. Cucumber Mosaic Virus-Encoded 2b Suppressor Inhibits Arabidopsis Argonaute1 Cleavage Activity to Counter Plant Defense. Genes Dev. 2006, 20, 3255–3268. [Google Scholar] [CrossRef]
- Chellappan, P.; Vanitharani, R.; Ogbe, F.; Fauquet, C.M. Effect of Temperature on Geminivirus-Induced RNA Silencing in Plants. Plant Physiol. 2005, 138, 1828–1841. [Google Scholar] [CrossRef]
- Covey, S.N.; Al-Kaff, N.S.; Lángara, A.; Turner, D.S. Plants Combat Infection by Gene Silencing. Nature 1997, 385, 781–782. [Google Scholar] [CrossRef]
- Wingard, S.A. Hosts and Symptoms of Ring Spot, A Virus Disease of Plants. J. Agric. Res. 1928, 37, 127–153. [Google Scholar]
- Xin, H.W.; Ding, S.W. Identification and Molecular Characterization of a Naturally Occurring RNA Virus Mutant Defective in the Initiation of Host Recovery. Virology 2003, 317, 253–262. [Google Scholar] [CrossRef]
- Jovel, J.; Walker, M.; Sanfaçon, H. Recovery of Nicotiana benthamiana Plants from a Necrotic Response Induced by a Nepovirus Is Associated with RNA Silencing but Not with Reduced Virus Titer. J. Virol. 2007, 81, 12285–12297. [Google Scholar] [CrossRef]
- Karran, R.A.; Sanfačon, H. Tomato Ringspot Virus Coat Protein Binds to ARGONAUTE 1 and Suppresses the Translation Repression of a Reporter Gene. Mol. Plant-Microbe Interact. 2014, 27, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.G.; MacFarlane, S.A.; Baulcombe, D.C. Gene Silencing without DNA: RNA-Mediated Cross-Protection between Viruses. Plant Cell 1999, 11, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Ciomperlik, J.J.; Omarov, R.T.; Scholthof, H.B. An Antiviral RISC Isolated from Tobacco Rattle Virus-Infected Plants. Virology 2011, 412, 117–124. [Google Scholar] [CrossRef]
- Kørner, C.J.; Pitzalis, N.; Peña, E.J.; Erhardt, M.; Vazquez, F.; Heinlein, M. Crosstalk between PTGS and TGS Pathways in Natural Antiviral Immunity and Disease Recovery. Nat. Plants 2018, 4, 157–164. [Google Scholar] [CrossRef]
- Kohm, B.A.; Goulden, M.G.; Gilbert, J.E.; Kavanagh, T.A.; Baulcombea, D.C. A Potato Virus X Resistance Gene Mediates an Induced, Nonspecific Resistance in Protoplasts. Plant Cell 1993, 5, 913–920. [Google Scholar] [CrossRef]
- Ritter, E.; Debener, T.; Barone, A.; Salamini, F.; Gebhardt, C. RFLP Mapping on Potato Chromosomes of Two Genes Controlling Extreme Resistance to Potato Virus X (PVX). Mol. Gen. Genet. 1991, 227, 81–85. [Google Scholar] [CrossRef]
- Fenyk, S.; Townsend, P.D.; Dixon, C.H.; Spies, G.B.; Campillo, A.D.S.E.; Slootweg, E.J.; Westerhof, L.B.; Gawehns, F.K.K.; Knight, M.R.; Sharples, G.J.; et al. The Potato Nucleotide-Binding Leucine-Rich Repeat (NLR) Immune Receptor Rx1 Is a Pathogen-Dependent DNA-Deforming Protein. J. Biol. Chem. 2015, 290, 24945–24960. [Google Scholar] [CrossRef]
- Goulden, M.G.; Baulcombe, D.C. Functionally Homologous Host Components Recognize Potato Virus X in Gomphrena globosa and Potato. Plant Cell 1993, 5, 921–930. [Google Scholar] [CrossRef]
- Knip, M.; Richard, M.M.S.; Oskam, L.; van Engelen, H.T.D.; Aalders, T.; Takken, F.L.W. Activation of Immune Receptor Rx1 Triggers Distinct Immune Responses Culminating in Cell Death after 4 Hours. Mol. Plant Pathol. 2019, 20, 575–588. [Google Scholar] [CrossRef]
- Slootweg, E.; Roosien, J.; Spiridon, L.N.; Petrescu, A.J.; Tameling, W.; Joosten, M.; Pomp, R.; van Schaik, C.; Dees, R.; Borst, J.W.; et al. Nucleocytoplasmic Distribution Is Required for Activation of Resistance by the Potato NB-LRR Receptor Rx1 and Is Balanced by Its Functional Domains. Plant Cell 2012, 22, 4195–4215. [Google Scholar] [CrossRef]
- Sukarta, O.C.A.; Diaz-Granados, A.; Slootweg, E.J.; Overmars, H.; van Schaik, C.; Pokhare, S.; Roosien, J.; Pomp, R.; Elashry, A.; Smant, G.; et al. Two Evolutionary Distinct Effectors from a Nematode and Virus Target RanGAP1 and 2 via the WPP Domain to Promote Disease. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hao, W.; Collier, S.M.; Moffett, P.; Chai, J. Structural Basis for the Interaction between the Potato Virus X Resistance Protein (Rx) and Its Cofactor Ran GTPase-Activating Protein 2 (RanGAP2). J. Biol. Chem. 2013, 288, 35868–35876. [Google Scholar] [CrossRef] [PubMed]
- Sacco, M.A.; Mansoor, S.; Moffett, P. A RanGAP Protein Physically Interacts with the NB-LRR Protein Rx, and Is Required for Rx-Mediated Viral Resistance. Plant J. 2007, 52, 82–93. [Google Scholar] [CrossRef]
- Tameling, W.I.L.; Baulcombe, D.C. Physical Association of the NB-LRR Resistance Protein Rx with a Ran GTPase–Activating Protein Is Required for Extreme Resistance to Potato Virus X. Plant Cell 2007, 19, 1682–1694. [Google Scholar] [CrossRef]
- Townsend, P.D.; Dixon, C.H.; Slootweg, E.J.; Sukarta, O.C.A.; Yang, A.W.H.; Hughes, T.R.; Sharples, G.J.; Pålsson, L.O.; Takken, F.L.W.; Goverse, A.; et al. The Intracellular Immune Receptor Rx1 Regulates the DNA-Binding Activity of a Golden2-like Transcription Factor. J. Biol. Chem. 2018, 293, 3218–3233. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.; Dinesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The Product of the Tobacco Mosaic Virus Resistance Gene N: Similarity to Toll and the Interleukin-1 Receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Erickson, F.L.; Holzberg, S.; Calderon-Urrea, A.; Handley, V.; Axtell, M.; Corr, C.; Baker, B. The Helicase Domain of the TMV Replicase Proteins Induces the N-Mediated Defence Response in Tobacco. Plant J. 1999, 18, 67–75. [Google Scholar] [CrossRef]
- Caplan, J.L.; Mamillapalli, P.; Burch-Smith, T.M.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplastic Protein NRIP1 Mediates Innate Immune Receptor Recognition of a Viral Effector. Cell 2008, 132, 449–462. [Google Scholar] [CrossRef]
- Burch-Smith, T.M.; Schiff, M.; Caplan, J.L.; Tsao, J.; Czymmek, K.; Dinesh-Kumar, S.P. A Novel Role for the TIR Domain in Association with Pathogen-Derived Elicitors. PLoS Biol. 2007, 5, e68. [Google Scholar] [CrossRef]
- Sakamoto, T.; Deguchi, M.; Brustolini, O.J.B.; Santos, A.A.; Silva, F.F.; Fontes, E.P.B. The Tomato RLK Superfamily: Phylogeny and Functional Predictions about the Role of the LRRII-RLK Subfamily in Antiviral Defense. BMC Plant Biol. 2012, 12, 229. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; He, P.; Li, J.; Heese, A.; Peck, S.C.; Nürnberger, T.; Martin, G.B.; Sheen, J. Bacterial Effectors Target the Common Signaling Partner BAK1 to Disrupt Multiple MAMP Receptor-Signaling Complexes and Impede Plant Immunity. Cell Host Microbe 2008, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Receptor-like Kinases from Arabidopsis Form a Monophyletic Gene Family Related to Animal Receptor Kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Santos, A.A.; Pires, S.R.; Rocha, C.S.; Saraiva, D.I.; Machado, J.P.B.; Mattos, E.C.; Fietto, L.G.; Fontes, E.P.B. Regulated Nuclear Trafficking of RpL10A Mediated by NIK1 Represents a Defense Strategy of Plant Cells against Virus. PLoS Pathog. 2008, 4, e1000247. [Google Scholar] [CrossRef] [PubMed]
- Berlanga, J.J.; Ventoso, I.; Harding, H.P.; Deng, J.; Ron, D.; Sonenberg, N.; Carrasco, L.; Haro, C.D. Antiviral Effect of the Mammalian Translation Initiation Factor 2α Kinase GCN2 against RNA Viruses. EMBO J. 2006, 25, 1730–1740. [Google Scholar] [CrossRef] [PubMed]
- Heinicke, L.A.; Wong, C.J.; Lary, J.; Nallagatla, S.R.; Diegelman-Parente, A.; Zheng, X.; Cole, J.L.; Bevilacqua, P.C. RNA Dimerization Promotes PKR Dimerization and Activation. J. Mol. Biol. 2009, 390, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Nallagatla, S.R.; Hwang, J.; Toroney, R.; Zheng, X.; Cameron, C.E.; Bevilacqua, P.C. 5′-Triphosphate-Dependent Activation of PKR by RNAs with Short Stem-Loops. Science 2007, 318, 1455–1458. [Google Scholar] [CrossRef]
- Toroney, R.; Nallagatla, S.R.; Boyer, J.A.; Cameron, C.E.; Bevilacqua, P.C. Regulation of PKR by HCV IRES RNA: Importance of Domain II and NS5A. J. Mol. Biol. 2010, 400, 393–412. [Google Scholar] [CrossRef]
- Qu, F.; Morris, T.J. Cap-Independent Translational Enhancement of Turnip Crinkle Virus Genomic and Subgenomic RNAs. J. Virol. 2000, 74, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.O.; Tomari, Y. Molecular Insights into MicroRNA-Mediated Translational Repression in Plants. Mol. Cell 2013, 52, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Lanet, E.; Delannoy, E.; Sormani, R.; Floris, M.; Brodersen, P.; Crété, P.; Voinnet, O.; Robaglia, C. Biochemical Evidence for Translational Repression by Arabidopsis MicroRNAs. Plant Cell 2009, 21, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. What Lava Lamps and Vinaigrette Can Teach Us about Cell Biology. Nature 2018, 555, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef]
- Buchan, J.R.; Parker, R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef]
- Xu, M.; Mazur, M.J.; Tao, X.; Kormelink, R. Cellular RNA Hubs: Friends and Foes of Plant Viruses. Mol. Plant. Microbe Interact. 2020, 33, 40–54. [Google Scholar] [CrossRef]
- Maldonado-Bonilla, L.D. Composition and Function of P Bodies in Arabidopsis thaliana. Front. Plant Sci. 2014, 5, 201. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Kroschwald, S.; Munder, M.C.; Maharana, S.; Franzmann, T.M.; Richter, D.; Ruer, M.; Hyman, A.A.; Alberti, S. Different Material States of Pub1 Condensates Define Distinct Modes of Stress Adaptation and Recovery. Cell Rep. 2018, 23, 3327–3339. [Google Scholar] [CrossRef]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-Body Formation Is a Consequence, Not the Cause, of RNA-Mediated Gene Silencing. Mol. Cell. Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Nover, L.; Fauth, M. Plant Stress Granules and MRNA Processing Bodies Are Distinct from Heat Stress Granules. Plant J. 2008, 56, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, J.Y.; Niu, Q.W.; Chua, N.H. Arabidopsis DCP2, DCP1, and VARICOSE Form a Decapping Complex Required for Postembryonic Development. Plant Cell 2006, 18, 3386–3398. [Google Scholar] [CrossRef]
- Sheth, U.; Parker, R. Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef]
- Beckham, C.J.; Light, H.R.; Nissan, T.A.; Ahlquist, P.; Parker, R.; Noueiry, A. Interactions between Brome Mosaic Virus RNAs and Cytoplasmic Processing Bodies. J. Virol. 2007, 81, 9759–9768. [Google Scholar] [CrossRef] [PubMed]
- Galão, R.P.; Chari, A.; Alves-Rodrigues, I.; Lobão, D.; Mas, A.; Kambach, C.; Fischer, U.; Díez, J. LSm1-7 Complexes Bind to Specific Sites in Viral RNA Genomes and Regulate Their Translation and Replication. RNA 2010, 16, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Motomura, K.; Le, Q.T.N.; Hamada, T.; Kutsuna, N.; Mano, S.; Nishimura, M.; Watanabe, Y. Diffuse Decapping Enzyme DCP2 Accumulates in DCP1 Foci Under Heat Stress in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 107–115. [Google Scholar] [CrossRef]
- Lageix, S.; Lanet, E.; Pouch-Pélissier, M.N.; Espagnol, M.C.; Robaglia, C.; Deragon, J.M.; Pélissier, T. Arabidopsis EIF2α Kinase GCN2 Is Essential for Growth in Stress Conditions and Is Activated by Wounding. BMC Plant Biol. 2008, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.J.W.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An Antiviral Defense Role of AGO2 in Plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.S.; Perry, K.L.; Moffett, P. ARGONAUTE2 Mediates RNA-Silencing Antiviral Defenses against Potato Virus X in Arabidopsis. Plant Physiol. 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.B.; Godon, C.; Mourrain, P.; Béclin, C.; Boutet, S.; Feuerbach, F.; Proux, F.; Vaucheret, H. Fertile Hypomorphic ARGONAUTE (Ago1) Mutants Impaired in Post-Transcriptional Gene Silencing and Virus Resistance. Plant Cell 2002, 14, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, H.B.; Alvarado, V.Y.; Vega-Arreguin, J.C.; Ciomperlik, J.; Odokonyero, D.; Brosseau, C.; Jaubert, M.; Zamora, A.; Moffett, P. Identification of an ARGONAUTE for Antiviral RNA Silencing in Nicotiana benthamiana. Plant Physiol. 2011, 156, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Ye, X.; Morris, T.J. Arabidopsis DRB4, AGO1, AGO7, and RDR6 Participate in a DCL4-Initiated Antiviral RNA Silencing Pathway Negatively Regulated by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 14732–14737. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 Is an RNA Slicer That Selectively Recruits MicroRNAs and Short Interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef]
- Yang, L.; Wu, G.; Poethig, R.S. Mutations in the GW-Repeat Protein SUO Reveal a Developmental Function for MicroRNA-Mediated Translational Repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 315–320. [Google Scholar] [CrossRef]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal Structure of Argonaute and Its Implications for RISC Slicer Activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to MiRNA Targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef]
- Karlowski, W.M.; Zielezinski, A.; Carrère, J.; Pontier, D.; Lagrange, T.; Cooke, R. Genome-Wide Computational Identification of WG/GW Argonaute-Binding Proteins in Arabidopsis. Nucleic Acids Res. 2010, 38, 4231–4245. [Google Scholar] [CrossRef]
- Tritschler, F.; Huntzinger, E.; Izaurralde, E. Role of GW182 Proteins and PABPC1 in the MiRNA Pathway: A Sense of Déjà Vu. Nat. Rev. Mol. Cell Biol. 2010, 11, 379–384. [Google Scholar] [CrossRef]
- Poulsen, C.; Vaucheret, H.; Brodersen, P. Lessons on RNA Silencing Mechanisms in Plants from Eukaryotic Argonaute Structures. Plant Cell 2013, 25, 22–37. [Google Scholar] [CrossRef]
- Azevedo, J.; Garcia, D.; Pontier, D.; Ohnesorge, S.; Yu, A.; Garcia, S.; Braun, L.; Bergdoll, M.; Hakimi, M.A.; Lagrange, T.; et al. Argonaute Quenching and Global Changes in Dicer Homeostasis Caused by a Pathogen-Encoded GW Repeat Protein. Genes Dev. 2010, 24, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Singh, J.; Li, D.; Qu, F. Temperature-Dependent Survival of Turnip Crinkle Virus-Infected Arabidopsis Plants Relies on an RNA Silencing-Based Defense That Requires DCL2, AGO2, and HEN1. J. Virol. 2012, 86, 6847–6854. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Tsai, C.H.; Lie, M.; Havecker, E.; Baulcombe, D.C.C. The Polerovirus Silencing Suppressor P0 Targets ARGONAUTE Proteins for Degradation. Curr. Biol. 2007, 17, 1609–1614. [Google Scholar] [CrossRef]
- Iki, T.; Tschopp, M.A.; Voinnet, O. Biochemical and Genetic Functional Dissection of the P38 Viral Suppressor of RNA Silencing. RNA 2017, 23, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.L.; Leh, V.; Lederer, C.; Maule, A.J. Turnip Crinkle Virus Coat Protein Mediates Suppression of RNA Silencing in Nicotiana benthamiana. Virology 2003, 306, 33–41. [Google Scholar] [CrossRef]
- Siddiqui, S.A.; Sarmiento, C.; Kiisma, M.; Koivumäki, S.; Lemmetty, A.; Truve, E.; Lehto, K. Effects of Viral Silencing Suppressors on Tobacco Ringspot Virus Infection in Two Nicotiana Species. J. Gen. Virol. 2008, 89, 1502–1508. [Google Scholar] [CrossRef]
- Chu, M.; Desvoyes, B.; Turina, M.; Noad, R.; Scholthof, H.B. Genetic Dissection of Tomato Bushy Stunt Virus P19-Protein-Mediated Host-Dependent Symptom Induction and Systemic Invasion. Virology 2000, 266, 79–87. [Google Scholar] [CrossRef]
- Lewsey, M.; Surette, M.; Robertson, F.C.; Ziebell, H.; Choi, S.H.; Ryu, K.H.; Canto, T.; Palukaitis, P.; Payne, T.; Walsh, J.A.; et al. The Role of the Cucumber Mosaic Virus 2b Protein in Viral Movement and Symptom Induction. Mol. Plant Pathol. 2009, 22, 642–654. [Google Scholar] [CrossRef]
- Lin, S.S.; Wu, H.W.; Jan, F.J.; Hou, R.F.; Yeh, S.D. Modifications of the Helper Component-Protease of Zucchini Yellow Mosaic Virus for Generation of Attenuated Mutants for Cross Protection Against Severe Infection. Phytopathology 2007, 97, 287–296. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, X.; Ding, S.-W. Viral Suppressors of RNA-Based Viral Immunity: Host Targets. Cell Host Microbe 2010, 8, 12–15. [Google Scholar] [CrossRef]
- Fernández-Calvino, L.; Martínez-Priego, L.; Szabo, E.Z.; Guzmán-Benito, I.; González, I.; Canto, T.; Lakatos, L.; Llave, C. Tobacco Rattle Virus 16K Silencing Suppressor Binds ARGONAUTE 4 and Inhibits Formation of RNA Silencing Complexes. J. Gen. Virol. 2016, 97, 246–257. [Google Scholar] [CrossRef]
- Chan, S.W.L.; Zilberman, D.; Xie, Z.; Johansen, L.K.; Carrington, J.C.; Jacobsen, S.E. RNA Silencing Genes Control de Novo DNA Methylation. Science 2004, 303, 1336. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, D.; Cao, X.; Jacobsen, S.E. ARGONAUTE4 Control of Locus-Specific SiRNA Accumulation and DNA and Histone Methylation. Science 2003, 299, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Oirdi, M.E.; Adurogbangba, A.; Ma, X.; Moffett, P. Antiviral Defense Involves AGO4 in an Arabidopsis-Potexvirus Interaction. Mol. Plant. Microbe Interact. 2016, 29, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, D.; Molnár, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyán, J. A Viral Protein Suppresses RNA Silencing and Binds Silencing-Generated, 21- to 25-Nucleotide Double-Stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef]
- Goeres, D.C.; Norman, J.M.V.; Zhang, W.; Fauver, N.A.; Spencer, M.L.; Sieburth, L.E. Components of the Arabidopsis MRNA Decapping Complex Are Required for Early Seedling Development. Plant Cell 2007, 19, 1549–1564. [Google Scholar] [CrossRef]
- Chicois, C.; Scheer, H.; Garcia, S.; Zuber, H.; Mutterer, J.; Chicher, J.; Hammann, P.; Gagliardi, D.; Garcia, D. The UPF1 Interactome Reveals Interaction Networks between RNA Degradation and Translation Repression Factors in Arabidopsis. Plant J. 2018, 96, 119–132. [Google Scholar] [CrossRef]
- Xu, J.; Chua, N.H. Arabidopsis Decapping 5 Is Required for MRNA Decapping, P-Body Formation, and Translational Repression during Postembryonic Development. Plant Cell 2009, 21, 3270–3279. [Google Scholar] [CrossRef]
- Bogamuwa, S.; Jang, J.-C. Plant Tandem CCCH Zinc Finger Proteins Interact with ABA, Drought, and Stress Response Regulators in Processing-Bodies and Stress Granules. PLoS ONE 2016, 11, e0151574. [Google Scholar] [CrossRef]
- Pomeranz, M.C.; Hah, C.; Lin, P.C.; Kang, S.G.; Finer, J.J.; Blackshear, P.J.; Jang, J.C. The Arabidopsis Tandem Zinc Finger Protein AtTZF1 Traffics between the Nucleus and Cytoplasmic Foci and Binds Both DNA and RNA. Plant Physiol. 2009, 152, 151–165. [Google Scholar] [CrossRef]
- Qu, J.; Kang, S.G.; Wang, W.; Musier-Forsyth, K.; Jang, J.C. The Arabidopsis thaliana Tandem Zinc Finger 1 (AtTZF1) Protein in RNA Binding and Decay. Plant J. 2014, 78, 452–467. [Google Scholar] [CrossRef]
- Chong, P.A.; Vernon, R.M.; Forman-Kay, J.D. RGG/RG Motif Regions in RNA Binding and Phase Separation. J. Mol. Biol. 2018, 430, 4650–4665. [Google Scholar] [CrossRef]
- Decker, C.J.; Teixeira, D.; Parker, R. Edc3p and a Glutamine/Asparagine-Rich Domain of Lsm4p Function in Processing Body Assembly in Saccharomyces Cerevisiae. J. Cell Biol. 2007, 179, 437–449. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.H.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The Disordered P Granule Protein LAF-1 Drives Phase Separation into Droplets with Tunable Viscosity and Dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Maruri-López, I.; Figueroa, N.E.; Hernández-Sánchez, I.E.; Chodasiewicz, M. Plant Stress Granules: Trends and Beyond. Front. Plant Sci. 2021, 12, 722643. [Google Scholar] [CrossRef]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604.e13. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch That Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e28. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of MRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Serino, G.; Deng, X.W.; Dinesh-Kumar, S.P. Role of SCF Ubiquitin-Ligase and the COP9 Signalosome in the N Gene–Mediated Resistance Response to Tobacco Mosaic Virus. Plant Cell 2002, 14, 1483–1496. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB Transcription Factors, COI1 and CTR1 in N-Mediated Resistance to Tobacco Mosaic Virus. Plant J. 2004, 38, 800–809. [Google Scholar] [CrossRef]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel Positive Regulatory Role for the SPL6 Transcription Factor in the N TIR-NB-LRR Receptor-Mediated Plant Innate Immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef]
- Pillai, R.S.; Artus, C.G.; Filipowicz, W. Tethering of Human Ago Proteins to mRNA Mimics the MiRNA-Mediated Repression of Protein Synthesis. RNA 2004, 10, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.P.S.; Han, L.; Arribas-Hernández, L.; Karelina, D.; Petersen, M.; Brodersen, P. Sensing of Viral RNA in Plants via a DICER-LIKE Ribonuclease. bioRxiv 2023. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Goubau, D.; Schlee, M.; Deddouche, S.; Pruijssers, A.J.; Zillinger, T.; Goldeck, M.; Schuberth, C.; Veen, A.G.V.D.; Fujimura, T.; Rehwinkel, J.; et al. Antiviral Immunity via RIG-I-Mediated Recognition of RNA Bearing 5′-Diphosphates. Nature 2014, 514, 372–375. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, S.; Jia, X.; Ge, Y.; Ling, T.; Nie, M.; Lan, X.; Chen, S.; Xu, A. Mitochondria-Localised ZNFX1 Functions as a DsRNA Sensor to Initiate Antiviral Responses through MAVS. Nat. Cell Biol. 2019, 21, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, N.; Yuan, B.; Weng, L.; Wang, F.; Liu, Y.J.; Zhang, Z. The Interaction between the Helicase DHX33 and IPS-1 as a Novel Pathway to Sense Double-Stranded RNA and RNA Viruses in Myeloid Dendritic Cells. Cell. Mol. Immunol 2013, 11, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Ikeda, M.; Matsumoto, M.; Watanabe, A.; Takeuchi, O.; Akira, S.; Kato, N.; Shimotohno, K.; Seya, T. Hepatitis C Virus Core Protein Abrogates the DDX3 Function That Enhances IPS-1-Mediated IFN–β Induction. PLoS ONE 2010, 5, e14258. [Google Scholar] [CrossRef]
- Langereis, M.A.; Feng, Q.; Kuppeveld, F.J. van MDA5 Localizes to Stress Granules, but This Localization Is Not Required for the Induction of Type I Interferon. J. Virol. 2013, 87, 6314–6325. [Google Scholar] [CrossRef] [PubMed]
- Onomoto, K.; Jogi, M.; Yoo, J.S.; Narita, R.; Morimoto, S.; Takemura, A.; Sambhara, S.; Kawaguchi, A.; Osari, S.; Nagata, K.; et al. Critical Role of an Antiviral Stress Granule Containing RIG-I and PKR in Viral Detection and Innate Immunity. PLoS ONE 2012, 7, e43031. [Google Scholar] [CrossRef]
- Slootweg, E.J.; Spiridon, L.N.; Martin, E.C.; Tameling, W.I.L.; Townsend, P.D.; Pomp, R.; Roosien, J.; Drawska, O.; Sukarta, O.C.A.; Schots, A.; et al. Distinct Roles of Non-Overlapping Surface Regions of the Coiled-Coil Domain in the Potato Immune Receptor Rx1. Plant Physiol. 2018, 178, 1310–1331. [Google Scholar] [CrossRef]
- Kosmacz, M.; Luzarowski, M.; Kerber, O.; Leniak, E.; Gutiérrez-Beltrán, E.; Moreno, J.C.; Gorka, M.; Szlachetko, J.; Veyel, D.; Graf, A.; et al. Interaction of 2′,3′-CAMP with Rbp47b Plays a Role in Stress Granule Formation. Plant Physiol. 2018, 177, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Song, W.; Tan, E.Y.J.; Liu, L.; Cao, Y.; Jirschitzka, J.; Li, E.; Logemann, E.; Xu, C.; Huang, S.; et al. TIR Domains of Plant Immune Receptors Are 2′,3′-CAMP/CGMP Synthetases Mediating Cell Death. Cell 2022, 185, 2370–2386.e18. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Mohr, I. Ribosome Biogenesis Restricts Innate Immune Responses to Virus Infection and DNA. eLife 2019, 8, e49551. [Google Scholar] [CrossRef] [PubMed]
- Hertz, M.I.; Landry, D.M.; Willis, A.E.; Luo, G.; Thompson, S.R. Ribosomal Protein S25 Dependency Reveals a Common Mechanism for Diverse Internal Ribosome Entry Sites and Ribosome Shunting. Mol. Cell. Biol. 2013, 33, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Y.; Su, W.C.; Jeng, K.S.; Chang, T.H.; Lai, M.M.C. Attenuation of 40S Ribosomal Subunit Abundance Differentially Affects Host and HCV Translation and Suppresses HCV Replication. PLoS Pathog. 2012, 8, e1002766. [Google Scholar] [CrossRef]
- Lee, A.S.Y.; Burdeinick-Kerr, R.; Whelan, S.P.J. A Ribosome-Specialized Translation Initiation Pathway Is Required for Cap-Dependent Translation of Vesicular Stomatitis Virus MRNAs. Proc. Natl. Acad. Sci. USA 2013, 110, 324–329. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, C.; Dittman, J.D.; Whitham, S.A. Differential Requirement of Ribosomal Protein S6 by Plant RNA Viruses with Different Translation Initiation Strategies. Virology 2009, 390, 163–173. [Google Scholar] [CrossRef]
- Bruns, A.N.; Li, S.; Mohannath, G.; Bisaro, D.M. Phosphorylation of Arabidopsis EIF4E and EIFiso4E by SnRK1 Inhibits Translation. FEBS J. 2019, 286, 3778. [Google Scholar] [CrossRef] [PubMed]
- McCready, K.; Spencer, V.; Kim, M. The Importance of TOR Kinase in Plant Development. Front. Plant Sci. 2020, 11, 16. [Google Scholar] [CrossRef]
- Soto-Burgos, J.; Bassham, D.C. SnRK1 Activates Autophagy via the TOR Signaling Pathway in Arabidopsis thaliana. PLoS ONE 2017, 12, e0182591. [Google Scholar] [CrossRef]
- Aznar, N.R.; Consolo, V.F.; Salerno, G.L.; Martínez-Noël, G.M.A. TOR Signaling Downregulation Increases Resistance to the Cereal Killer Fusarium Graminearum. Plant Signal. Behav. 2018, 13, e1414120. [Google Scholar] [CrossRef] [PubMed]
- Ouibrahim, L.; Rubio, A.G.; Moretti, A.; Montané, M.-H.; Menand, B.; Meyer, C.; Robaglia, C.; Caranta, C. Potyviruses Differ in Their Requirement for TOR Signalling. J. Gen. Virol. 2015, 96, 2898–2903. [Google Scholar] [CrossRef] [PubMed]
- Marash, I.; Leibman-Markus, M.; Gupta, R.; Avni, A.; Bar, M. TOR Inhibition Primes Immunity and Pathogen Resistance in Tomato in a Salicylic Acid-Dependent Manner. Mol. Plant Pathol. 2022, 23, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vermeulen, A.; Takken, F.L.W.; Sánchez-Camargo, V.A. Translation Arrest: A Key Player in Plant Antiviral Response. Genes 2023, 14, 1293. https://doi.org/10.3390/genes14061293
Vermeulen A, Takken FLW, Sánchez-Camargo VA. Translation Arrest: A Key Player in Plant Antiviral Response. Genes. 2023; 14(6):1293. https://doi.org/10.3390/genes14061293
Chicago/Turabian StyleVermeulen, Annemarie, Frank L. W. Takken, and Victor A. Sánchez-Camargo. 2023. "Translation Arrest: A Key Player in Plant Antiviral Response" Genes 14, no. 6: 1293. https://doi.org/10.3390/genes14061293
APA StyleVermeulen, A., Takken, F. L. W., & Sánchez-Camargo, V. A. (2023). Translation Arrest: A Key Player in Plant Antiviral Response. Genes, 14(6), 1293. https://doi.org/10.3390/genes14061293








