Determination of Effect Sizes for Power Analysis for Microbiome Studies Using Large Microbiome Databases
Abstract
1. Introduction
2. Materials and Methods
2.1. Statistical Methodology
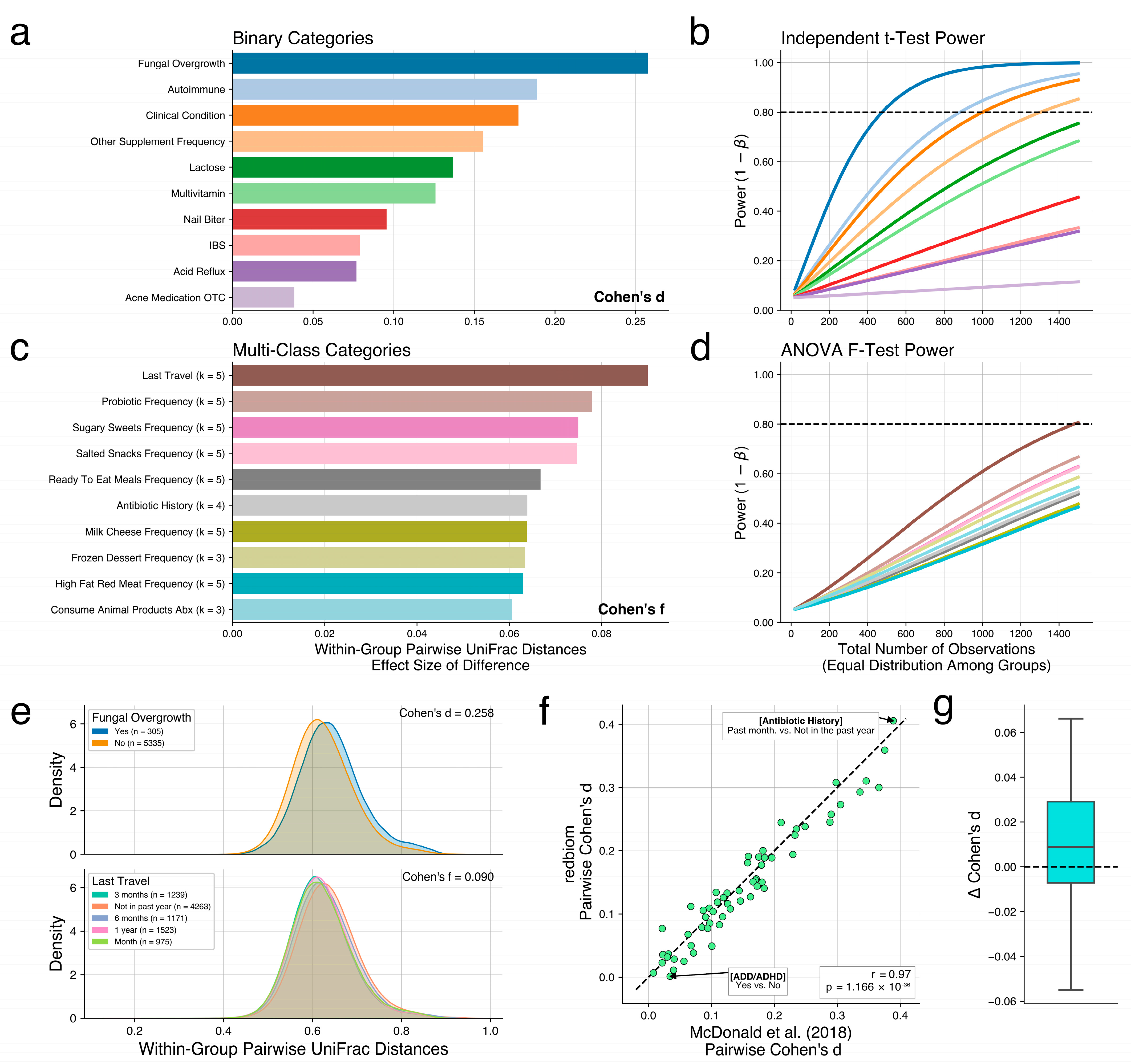
2.2. Interactive Exploration of Community Differences
2.3. Analysis of AGP Data
2.4. Analysis of Study of Latinos Data
3. Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Code Availability
References
- Sullivan, G.M.; Feinn, R. Using Effect Size—Or Why the P Value Is Not Enough. J. Grad. Med. Educ. 2012, 4, 279–282. [Google Scholar] [CrossRef]
- Baguley, T. Standardized or simple effect size: What should be reported? Br. J. Psychol. 2009, 100, 603–617. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis. Curr. Dir. Psychol. Sci. 1992, 1, 98–101. [Google Scholar] [CrossRef]
- McDonald, D.; Hyde, E.; Debelius, J.W.; Morton, J.T.; Gonzalez, A.; Ackermann, G.; Alexander, A. American Gut: An Open Platform for Citizen Science Microbiome Research. mSystems 2018, 3, e00031-18. [Google Scholar] [CrossRef]
- TEDDY Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann. N. Y. Acad. Sci. 2008, 1150, 1–13. [Google Scholar] [CrossRef]
- Vartiainen, E.; Jousilahti, P.; Alfthan, G.; Sundvall, J.; Pietinen, P.; Puska, P. Cardiovascular risk factor changes in Finland, 1972–1997. Int. J. Epidemiol. 2000, 29, 49–56. [Google Scholar] [CrossRef]
- Casals-Pascual, C.; González, A.; Vázquez-Baeza, Y.; Song, S.J.; Jiang, L.; Knight, R. Microbial Diversity in Clinical Microbiome Studies: Sample Size and Statistical Power Considerations. Gastroenterology 2020, 158, 1524–1528. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McClorry, S.; Zavaleta, N.; Llanos, A.; Casapía, M.; Lönnerdal, B.; Slupsky, C.M. Anemia in infancy is associated with alterations in systemic metabolism and microbial structure and function in a sex-specific manner: An observational study. Am. J. Clin. Nutr. 2018, 108, 1238–1248. [Google Scholar] [CrossRef]
- Yang, L.; Chen, J. A comprehensive evaluation of microbial differential abundance analysis methods: Current status and potential solutions. Microbiome 2022, 10, 130. [Google Scholar] [CrossRef]
- Dwiyanto, J.; Hussain, M.H.; Reidpath, D.; Ong, K.S.; Qasim, A.; Lee, S.W.H.; Lee, S.M.; Foo, S.C.; Chong, C.W.; Rahman, S. Ethnicity influences the gut microbiota of individuals sharing a geographical location: A cross-sectional study from a middle-income country. Sci. Rep. 2021, 11, 2618. [Google Scholar] [CrossRef]
- Park, J.; Kato, K.; Murakami, H.; Hosomi, K.; Tanisawa, K.; Nakagata, T.; Ohno, H.; Konishi, K.; Kawashima, H.; Chen, Y.-A.; et al. Comprehensive analysis of gut microbiota of a healthy population and covariates affecting microbial variation in two large Japanese cohorts. BMC Microbiol. 2021, 21, 151. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Lawrence Erlbaum Associates: Mahwah, NJ, USA, 1988; pp. 274–275. [Google Scholar]
- Gonzalez, A.; Navas-Molina, J.A.; Kosciolek, T.; McDonald, D.; Vázquez-Baeza, Y.; Ackermann, G.; Dereus, J.; Janssen, S.; Swafford, A.D.; Orchanian, S.B.; et al. Qiita: Rapid, web-enabled microbiome meta-analysis. Nat. Methods 2018, 15, 796–798. [Google Scholar] [CrossRef]
- McDonald, D.; Kaehler, B.; Gonzalez, A.; DeReus, J.; Ackermann, G.; Marotz, C.; Huttley, G.; Knight, R. redbiom: A Rapid Sample Discovery and Feature Characterization System. mSystems 2019, 4, e00215-19. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Debelius, J.; Morton, J.T.; Hyde, E.; Robbins-Pianka, A.; Knight, R. Correcting for Microbial Blooms in Fecal Samples during Room-Temperature Shipping. mSystems 2017, 2, e00199-16. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Mirarab, S.; Nguyen, N.; Warnow, T. SEPP: SATé-enabled phylogenetic placement. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2012, 2011, 247–258. [Google Scholar] [CrossRef]
- McDonald, D.; Vázquez-Baeza, Y.; Koslicki, D.; McClelland, J.; Reeve, N.; Xu, Z.; Gonzalez, A.; Knight, R. Striped UniFrac: Enabling microbiome analysis at unprecedented scale. Nat. Methods 2018, 15, 847–848. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Morton, J.T.; Marotz, C.; Washburne, A.; Silverman, J.; Zaramela, L.S.; Edlund, A.; Zengler, K.; Knight, R. Establishing microbial composition measurement standards with reference frames. Nat. Commun. 2019, 10, 2719. [Google Scholar] [CrossRef]
- Fedarko, M.W.; Martino, C.; Morton, J.T.; González, A.; Rahman, G.; A Marotz, C.; Minich, J.J.; E Allen, E.; Knight, R. Visualizing’omic feature rankings and log-ratios using Qurro. NAR Genom. Bioinform. 2020, 2, lqaa023. [Google Scholar] [CrossRef]
- Kaplan, R.C.; Wang, Z.; Usyk, M.; Sotres-Alvarez, D.; Daviglus, M.L.; Schneiderman, N.; Talavera, G.A.; Gellman, M.D.; Thyagarajan, B.; Moon, J.-Y.; et al. Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 2019, 20, 219. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, G.; McDonald, D.; Gonzalez, A.; Vázquez-Baeza, Y.; Jiang, L.; Casals-Pascual, C.; Hakim, D.; Dilmore, A.H.; Nowinski, B.; Peddada, S.; et al. Determination of Effect Sizes for Power Analysis for Microbiome Studies Using Large Microbiome Databases. Genes 2023, 14, 1239. https://doi.org/10.3390/genes14061239
Rahman G, McDonald D, Gonzalez A, Vázquez-Baeza Y, Jiang L, Casals-Pascual C, Hakim D, Dilmore AH, Nowinski B, Peddada S, et al. Determination of Effect Sizes for Power Analysis for Microbiome Studies Using Large Microbiome Databases. Genes. 2023; 14(6):1239. https://doi.org/10.3390/genes14061239
Chicago/Turabian StyleRahman, Gibraan, Daniel McDonald, Antonio Gonzalez, Yoshiki Vázquez-Baeza, Lingjing Jiang, Climent Casals-Pascual, Daniel Hakim, Amanda Hazel Dilmore, Brent Nowinski, Shyamal Peddada, and et al. 2023. "Determination of Effect Sizes for Power Analysis for Microbiome Studies Using Large Microbiome Databases" Genes 14, no. 6: 1239. https://doi.org/10.3390/genes14061239
APA StyleRahman, G., McDonald, D., Gonzalez, A., Vázquez-Baeza, Y., Jiang, L., Casals-Pascual, C., Hakim, D., Dilmore, A. H., Nowinski, B., Peddada, S., & Knight, R. (2023). Determination of Effect Sizes for Power Analysis for Microbiome Studies Using Large Microbiome Databases. Genes, 14(6), 1239. https://doi.org/10.3390/genes14061239





