Cholecalciferol Supplementation Induced Up-Regulation of SARAF Gene and Down-Regulated miR-155-5p Expression in Slovenian Patients with Multiple Sclerosis
Abstract
1. Introduction
- Measure the miR-155-5p expression in peripheral blood mononuclear cells (PBMCs);
- Profile the genome for variants associated with cholecalciferol uptake;
- Stringently select genetic targets of miR-155-5p and extract genetic variants associated with cholecalciferol pathways;
- Calculate and assess expression quantitative trait loci (eQTLs) between aforementioned genetic variants, miR-155-5p, and miR-155-5p target genes;
- Identify target genes where both eQTLs are observed and measure the expression of the corresponding gene.
2. Materials and Methods
2.1. Subjects
2.2. Sample Collection
2.3. DNA, mRNA, and miRNA Extraction
2.4. MiR-155-5p RT-qPCR
2.5. Genotyping, Imputation, and Association Analysis
2.6. Integration of Genomics to miRNA-155-5p Targets
2.7. RT-qPCR Target Gene Validation
2.8. Statistical Analyses
3. Results
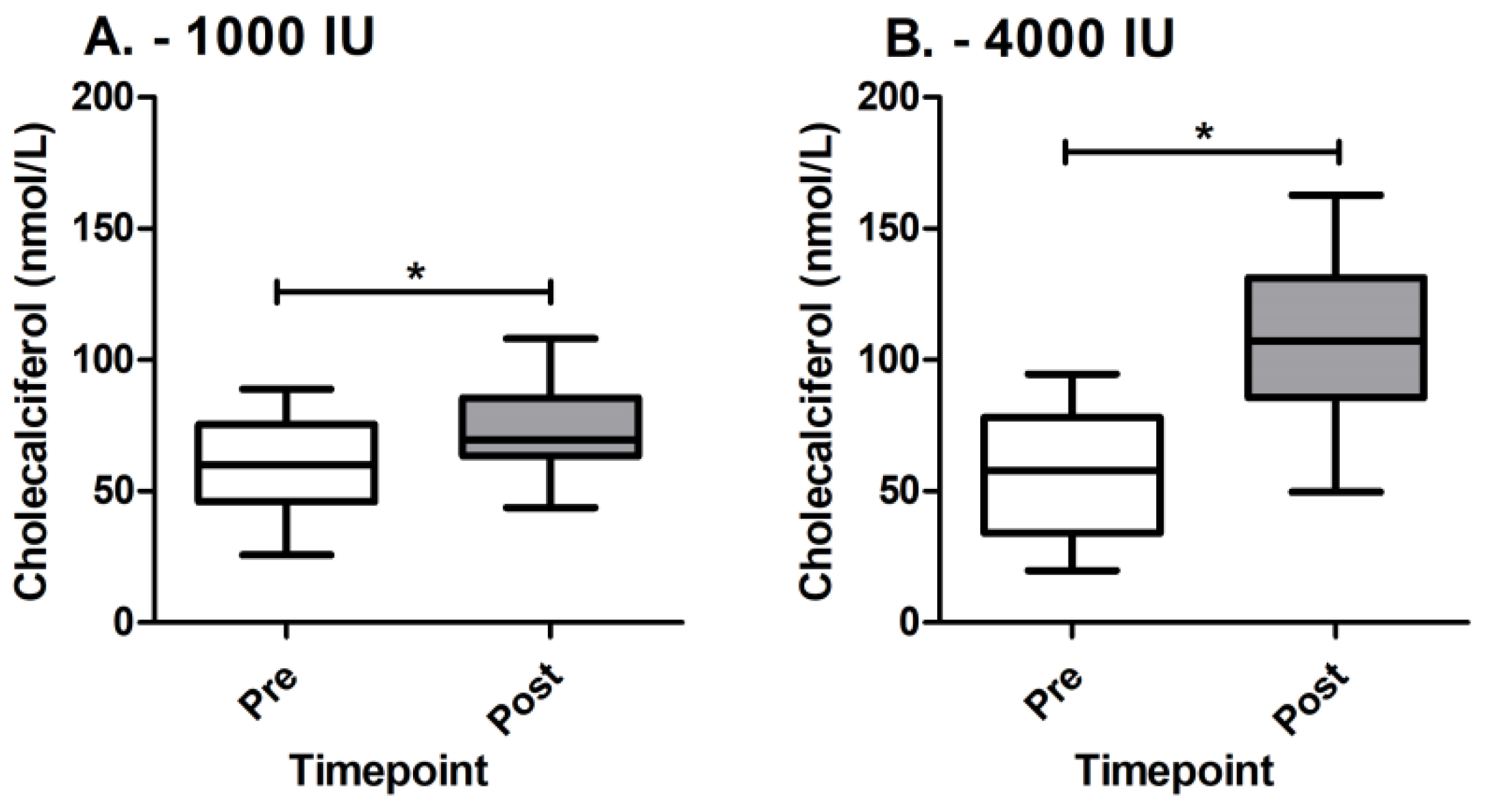
3.1. Estimation of Cholecalciferol Supplementation
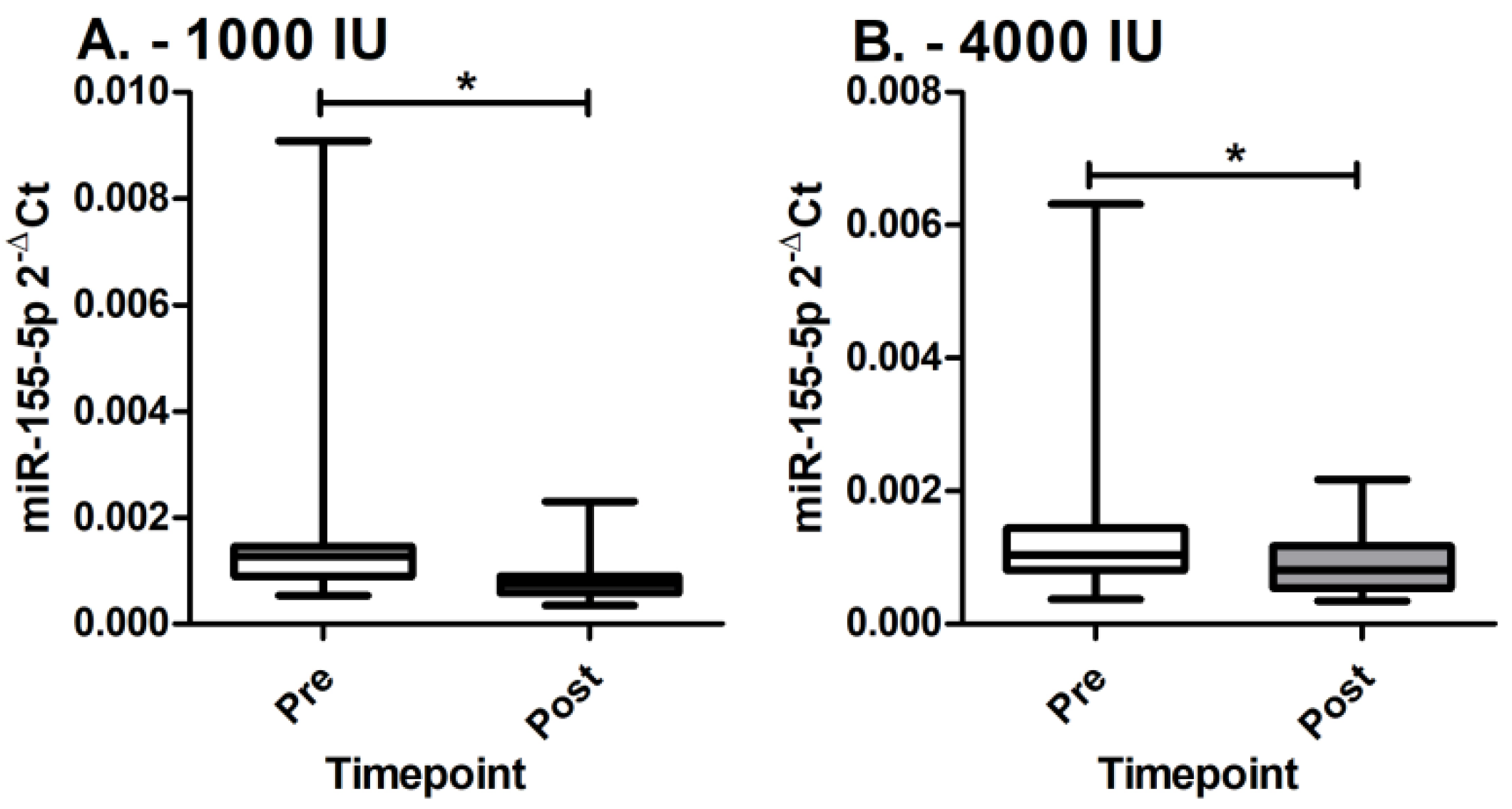
3.2. MiR-155-5p Expression
3.3. MiR-155-5p Targets, Integration to Genomics, and eQTL Estimation
3.4. Target Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramagopalan, S.V.; Sadovnick, A.D. Epidemiology of multiple sclerosis. Neurol. Clin. 2011, 29, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Nylander, A.; Hafler, D.A. Multiple sclerosis. J. Clin. Investig. 2012, 122, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Karakatič, S.; Magdič, J.; Karakatič, S.; Omerzu, T.; Modrič, E.; Hojs Fabjan, T. Diagnostic relevance of free light chain indices and their relation to the clinical presentation of multiple sclerosis. Acta Med. Biotech. 2020, 13, 23–32. [Google Scholar] [CrossRef]
- Popescu, B.F.; Pirko, I.; Lucchinetti, C.F. Pathology of multiple sclerosis: Where do we stand? Continuum 2013, 19, 901–921. [Google Scholar] [CrossRef]
- Mokry, L.E.; Ross, S.; Ahmad, O.S.; Forgetta, V.; Smith, G.D.; Goltzman, D.; Leong, A.; Greenwood, C.M.; Thanassoulis, G.; Richards, J.B. Vitamin D and Risk of Multiple Sclerosis: A Mendelian Randomization Study. PLoS Med. 2015, 12, e1001866. [Google Scholar] [CrossRef]
- Salzer, J.; Hallmans, G.; Nystrom, M.; Stenlund, H.; Wadell, G.; Sundstrom, P. Vitamin D as a protective factor in multiple sclerosis. Neurology 2012, 79, 2140–2145. [Google Scholar] [CrossRef]
- Manousaki, D.; Mitchell, R.; Dudding, T.; Haworth, S.; Harroud, A.; Forgetta, V.; Shah, R.L.; Luan, J.; Langenberg, C.; Timpson, N.J.; et al. Genome-wide Association Study for Vitamin D Levels Reveals 69 Independent Loci. Am. J. Hum. Genet. 2020, 106, 327–337. [Google Scholar] [CrossRef]
- Lucas, R.M.; Ponsonby, A.L.; Dear, K.; Valery, P.C.; Pender, M.P.; Taylor, B.V.; Kilpatrick, T.J.; Dwyer, T.; Coulthard, A.; Chapman, C.; et al. Sun exposure and vitamin D are independent risk factors for CNS demyelination. Neurology 2011, 76, 540–548. [Google Scholar] [CrossRef]
- Ramagopalan, S.V.; Handel, A.E.; Giovannoni, G.; Rutherford Siegel, S.; Ebers, G.C.; Chaplin, G. Relationship of UV exposure to prevalence of multiple sclerosis in England. Neurology 2011, 76, 1410–1414. [Google Scholar] [CrossRef]
- Simpson, S., Jr.; Wang, W.; Otahal, P.; Blizzard, L.; van der Mei, I.A.F.; Taylor, B.V. Latitude continues to be significantly associated with the prevalence of multiple sclerosis: An updated meta-analysis. J. Neurol. Neurosurg. Psychiatry 2019, 90, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Scazzone, C.; Agnello, L.; Bivona, G.; Lo Sasso, B.; Ciaccio, M. Vitamin D and Genetic Susceptibility to Multiple Sclerosis. Biochem. Genet. 2021, 59, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Sondergaard, H.B.; Hesse, D.; Krakauer, M.; Sorensen, P.S.; Sellebjerg, F. Differential microRNA expression in blood in multiple sclerosis. Mult. Scler. 2013, 19, 1849–1857. [Google Scholar] [CrossRef] [PubMed]
- Mahboobi, R.; Fallah, F.; Yadegar, A.; Dara, N.; Kazemi Aghdam, M.; Asgari, B.; Hakemi-Vala, M. Expression analysis of miRNA-155 level in Helicobacter pylori related inflammation and chronic gastritis. Iran J. Microbiol. 2022, 14, 495–502. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Maciak, K.; Dziedzic, A.; Miller, E.; Saluk-Bijak, J. miR-155 as an Important Regulator of Multiple Sclerosis Pathogenesis. A Review. Int. J. Mol. Sci. 2021, 22, 4332. [Google Scholar] [CrossRef]
- Asadpour-Behzadi, A.; Kariminik, A.; Kheirkhah, B. MicroRNA-155 is a main part of proinflammatory puzzle during severe coronavirus disease 2019 (COVID-19). Allergol. Immunopathol. 2023, 51, 115–119. [Google Scholar] [CrossRef]
- Rastegar-Moghaddam, S.H.; Ebrahimzadeh-Bideskan, A.; Shahba, S.; Malvandi, A.M.; Mohammadipour, A. Roles of the miR-155 in Neuroinflammation and Neurological Disorders: A Potent Biological and Therapeutic Target. Cell. Mol. Neurobiol. 2023, 43, 455–467. [Google Scholar] [CrossRef]
- McCoy, C.E. miR-155 Dysregulation and Therapeutic Intervention in Multiple Sclerosis. Adv. Exp. Med. Biol. 2017, 1024, 111–131. [Google Scholar] [CrossRef]
- Paraboschi, E.M.; Solda, G.; Gemmati, D.; Orioli, E.; Zeri, G.; Benedetti, M.D.; Salviati, A.; Barizzone, N.; Leone, M.; Duga, S.; et al. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int. J. Mol. Sci. 2011, 12, 8695–8712. [Google Scholar] [CrossRef]
- Lopez-Ramirez, M.A.; Wu, D.; Pryce, G.; Simpson, J.E.; Reijerkerk, A.; King-Robson, J.; Kay, O.; de Vries, H.E.; Hirst, M.C.; Sharrack, B.; et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J. 2014, 28, 2551–2565. [Google Scholar] [CrossRef] [PubMed]
- Ali Ashrafi, S.; Asadi, M.; Shanehbandi, D.; Sadigh Eteghad, S.; Fazlollahi, A.; Nejadghaderi, S.A.; Shaafi, S. Association between miRNA-145 and miRNA-155 expression in peripheral blood mononuclear cells of patients with multiple sclerosis: A case-control study. BMC Neurol. 2022, 22, 405. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, M.; Lenhof, H.P.; Ruprecht, K.; Meese, E. Multiple sclerosis: MicroRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE 2009, 4, e7440. [Google Scholar] [CrossRef]
- Li, Y.C.; Chen, Y.; Liu, W.; Thadhani, R. MicroRNA-mediated mechanism of vitamin D regulation of innate immune response. J. Steroid. Biochem. Mol. Biol. 2014, 144 Pt A, 81–86. [Google Scholar] [CrossRef]
- Hanwell, H.E.; Vieth, R.; Cole, D.E.; Scillitani, A.; Modoni, S.; Frusciante, V.; Ritrovato, G.; Chiodini, I.; Minisola, S.; Carnevale, V. Sun exposure questionnaire predicts circulating 25-hydroxyvitamin D concentrations in Caucasian hospital workers in southern Italy. J. Steroid Biochem. Mol. Biol. 2010, 121, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, L.; Brekke, H.K.; Brembeck, P.; Augustin, H. A Short Questionnaire for Assessment of Dietary Vitamin D Intake. Eur. J. Nutr. Food Saf. 2014, 4, 150–156. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010, 5, 1564–1573. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schonherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Boerwinkle, E.; Xiong, M. Epistasis analysis for quantitative traits by functional regression model. Genome Res. 2014, 24, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.B.; Chiu, C.M.; Hsu, S.D.; Huang, W.Y.; Chien, C.H.; Lee, T.Y.; Huang, H.D. miRTar: An integrated system for identifying miRNA-target interactions in human. BMC Bioinform. 2011, 12, 300. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Gorenjak, M.; Repnik, K.; Jezernik, G.; Jurgec, S.; Skok, P.; Potocnik, U. Genetic prediction profile for adalimumab response in Slovenian Crohn’s disease patients. Z Gastroenterol. 2019, 57, 1218–1225. [Google Scholar] [CrossRef]
- Consortium, G. Erratum: Genetic effects on gene expression across human tissues. Nature 2018, 553, 530. [Google Scholar] [CrossRef]
- Tonacci, A.; Bagnato, G.; Pandolfo, G.; Billeci, L.; Sansone, F.; Conte, R.; Gangemi, S. MicroRNA Cross-Involvement in Autism Spectrum Disorders and Atopic Dermatitis: A Literature Review. J. Clin. Med. 2019, 8, 88. [Google Scholar] [CrossRef]
- Aljawadi, Z.A.; Al-Derzi, A.R.; Abdul-Majeed, B.A.; Almahdawi, A.M. MicroRNAs (20a, 146a, 155, and 145) expressions in a sample of Iraqi patients with multiple sclerosis. J. Fac. Med. Baghdad 2016, 58, 371–377. [Google Scholar] [CrossRef]
- Hu, X.; Li, M.; Zhang, Y.; Sang, K.; Li, W.; Liu, B.; Wan, L.; Du, B.; Qian, J.; Meng, F.; et al. An innovative immunotherapeutic strategy for rheumatoid arthritis: Selectively suppressing angiogenesis and osteoclast differentiation by fully human antibody targeting thymocyte antigen-1. Exp. Cell. Res. 2023, 424, 113490. [Google Scholar] [CrossRef]
- Mithal, A.; Wahl, D.A.; Bonjour, J.P.; Burckhardt, P.; Dawson-Hughes, B.; Eisman, J.A.; El-Hajj Fuleihan, G.; Josse, R.G.; Lips, P.; Morales-Torres, J. Global vitamin D status and determinants of hypovitaminosis D. Osteoporos. Int. 2009, 20, 1807–1820. [Google Scholar] [CrossRef] [PubMed]
- Zomot, E.; Achildiev Cohen, H.; Dagan, I.; Militsin, R.; Palty, R. Bidirectional regulation of calcium release-activated calcium (CRAC) channel by SARAF. J. Cell. Biol. 2021, 220, e202104007. [Google Scholar] [CrossRef]
- Taha, S.; Aljishi, M.; Alsharoqi, I.; Bakhiet, M. Differential upregulation of the hypothetical transmembrane protein 66 (TMEM66) in multiple sclerosis patients with potential inflammatory response. Biomed. Rep. 2015, 3, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Macian, F. NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 2005, 5, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Deb, J.; Patra, A.K.; Thuy Pham, D.A.; Chen, W.; Vaeth, M.; Berberich-Siebelt, F.; Klein-Hessling, S.; Lamperti, E.D.; Reifenberg, K.; et al. NFATc1 affects mouse splenic B cell function by controlling the calcineurin-NFAT signaling network. J. Exp. Med. 2011, 208, 823–839. [Google Scholar] [CrossRef]
- Vaeth, M.; Muller, G.; Stauss, D.; Dietz, L.; Klein-Hessling, S.; Serfling, E.; Lipp, M.; Berberich, I.; Berberich-Siebelt, F. Follicular regulatory T cells control humoral autoimmunity via NFAT2-regulated CXCR5 expression. J. Exp. Med. 2014, 211, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Vaeth, M.; Feske, S. NFAT control of immune function: New Frontiers for an Abiding Trooper. F1000Research 2018, 7, 260. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, W.; Sun, T.; Huang, Y.; Wang, Y.; Deb, D.K.; Yoon, D.; Kong, J.; Thadhani, R.; Li, Y.C. 1,25-Dihydroxyvitamin D promotes negative feedback regulation of TLR signaling via targeting microRNA-155-SOCS1 in macrophages. J. Immunol. 2013, 190, 3687–3695. [Google Scholar] [CrossRef]
- Saridas, F.; Tezcan Unlu, H.; Cecener, G.; Egeli, U.; Sabour Takanlou, M.; Sabour Takanlou, L.; Tunca, B.; Zarifoglu, M.; Turan, O.F.; Taskapilioglu, O. The expression and prognostic value of miR-146a and miR-155 in Turkish patients with multiple sclerosis. Neurol. Res. 2022, 44, 217–223. [Google Scholar] [CrossRef]


| 1000 IU | 4000 IU | p Value | |
|---|---|---|---|
| Sex (M/F) | 11/23 | 11/20 | 0.800 |
| Age (years) | 39.7 ± 9.5 | 42.2 ± 9.2 | 0.261 |
| MS duration (months) | 9.3 ± 4.7 | 10.7 ± 6.4 | 0.485 |
| EDSS | 2.0 ± 1.6 | 2.3 ± 1.4 | 0.437 |
| MSFC | 0.4 ± 0.4 | 0.2 ± 0.6 | 0.451 |
| Cholecalciferol | 59.3 ± 18.0 | 56.2 ± 22.0 | 0.650 |
| Parathyroid hormone | 41.8 ± 19.3 | 46.0 ± 16.8 | 0.147 |
| Creatinine | 62.4 ± 11.9 | 66.5 ± 14.8 | 0.269 |
| Calcium | 1.2 ± 0.2 | 1.2 ± 0.2 | 0.230 |
| Phosphate | 1.0 ± 0.2 | 1.0 ± 0.2 | 0.916 |
| CRP | 3.7 ± 2.2 | 4.5 ± 3.5 | 0.137 |
| Variant | Gene | Location | p Value |
|---|---|---|---|
| rs2271367 | SARAF | Chr8:29923732 | 0.024 |
| rs74849864 | TCF4 | Chr18:52924695 | 0.022 |
| rs62129063 | SMARCA4 | Chr19:11136006 | 0.048 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gselman, S.; Fabjan, T.H.; Bizjak, A.; Potočnik, U.; Gorenjak, M. Cholecalciferol Supplementation Induced Up-Regulation of SARAF Gene and Down-Regulated miR-155-5p Expression in Slovenian Patients with Multiple Sclerosis. Genes 2023, 14, 1237. https://doi.org/10.3390/genes14061237
Gselman S, Fabjan TH, Bizjak A, Potočnik U, Gorenjak M. Cholecalciferol Supplementation Induced Up-Regulation of SARAF Gene and Down-Regulated miR-155-5p Expression in Slovenian Patients with Multiple Sclerosis. Genes. 2023; 14(6):1237. https://doi.org/10.3390/genes14061237
Chicago/Turabian StyleGselman, Saša, Tanja Hojs Fabjan, Anja Bizjak, Uroš Potočnik, and Mario Gorenjak. 2023. "Cholecalciferol Supplementation Induced Up-Regulation of SARAF Gene and Down-Regulated miR-155-5p Expression in Slovenian Patients with Multiple Sclerosis" Genes 14, no. 6: 1237. https://doi.org/10.3390/genes14061237
APA StyleGselman, S., Fabjan, T. H., Bizjak, A., Potočnik, U., & Gorenjak, M. (2023). Cholecalciferol Supplementation Induced Up-Regulation of SARAF Gene and Down-Regulated miR-155-5p Expression in Slovenian Patients with Multiple Sclerosis. Genes, 14(6), 1237. https://doi.org/10.3390/genes14061237







