Efficiency of Biological Typing Methods in Maize Hybrid Genetic Purity Estimation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Varietal Uniformity Estimation by Morphological Markers
2.3. Genetic Purity Estimation by Protein Markers
2.4. Genetic Purity Estimation by Molecular Markers
3. Results
3.1. Varietal Uniformity Estimation by Morphological Markers
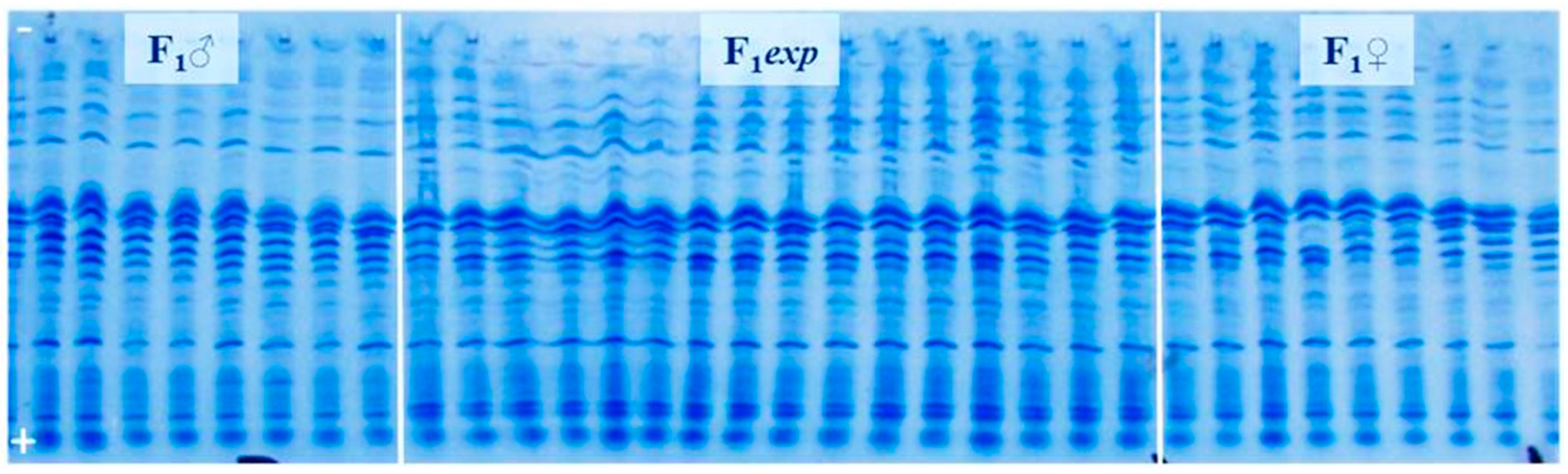
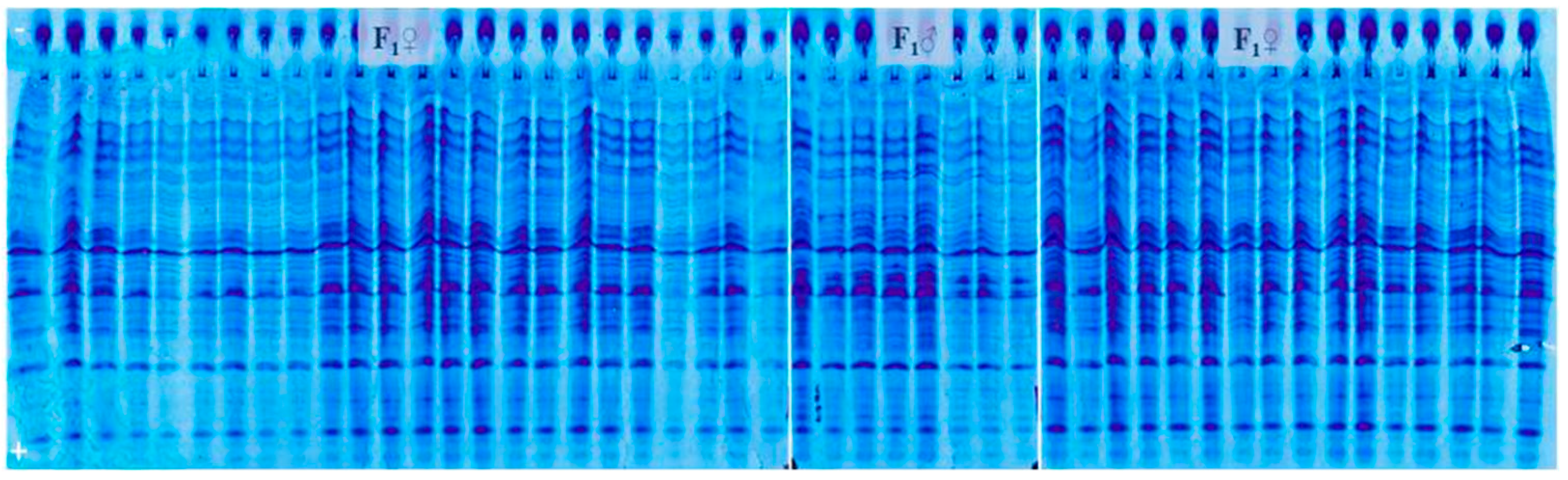
3.2. Genetic Purity Estimation by Protein Markers
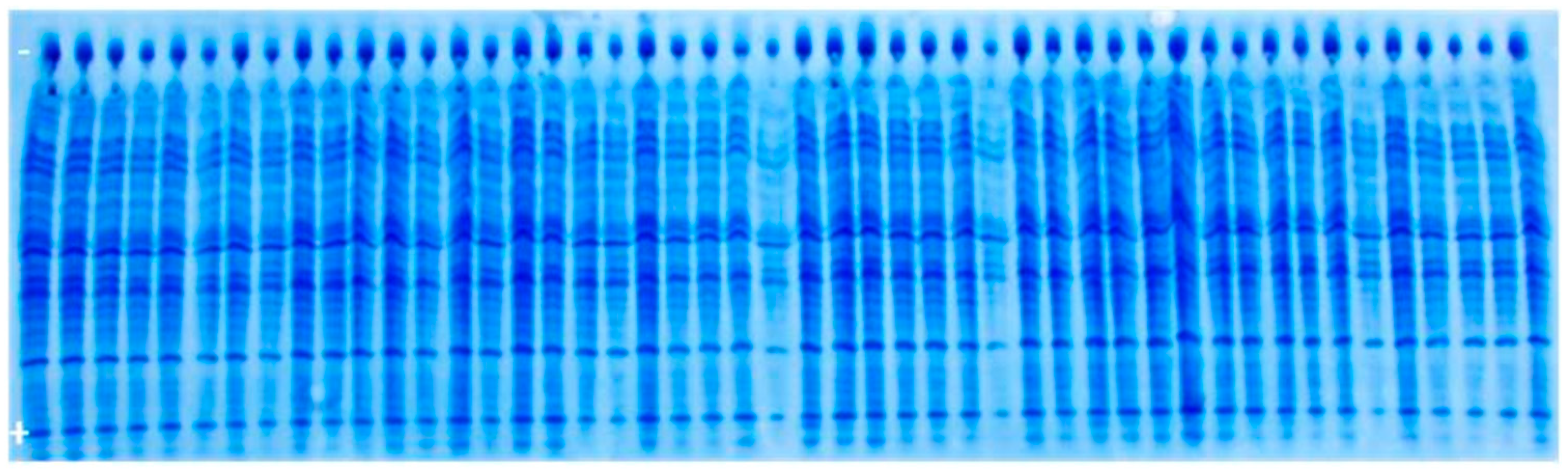
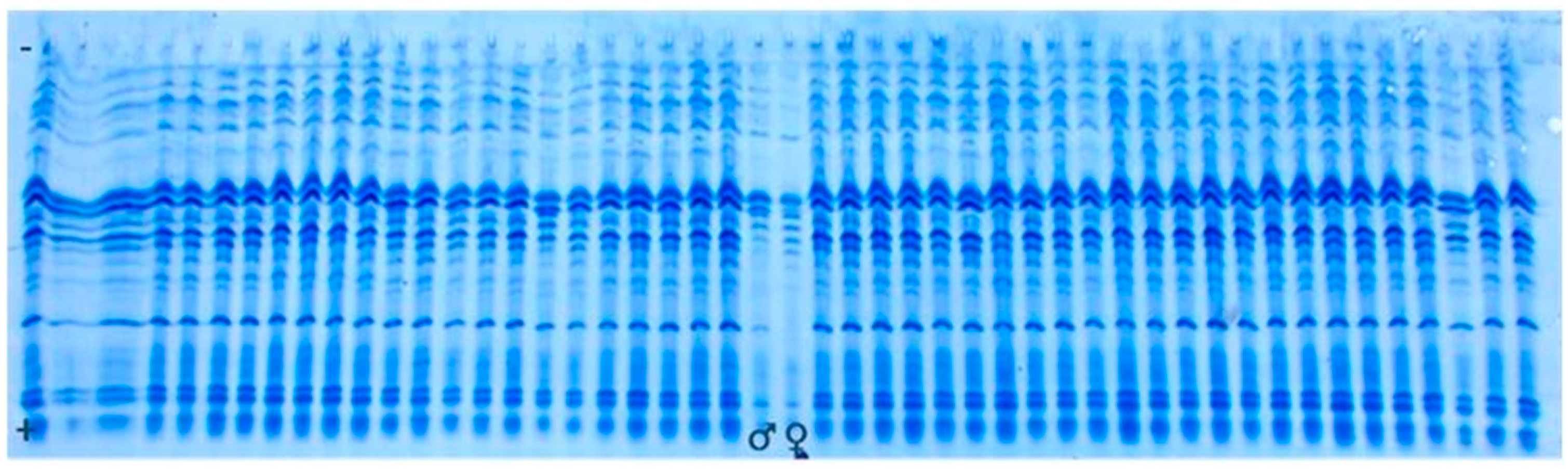
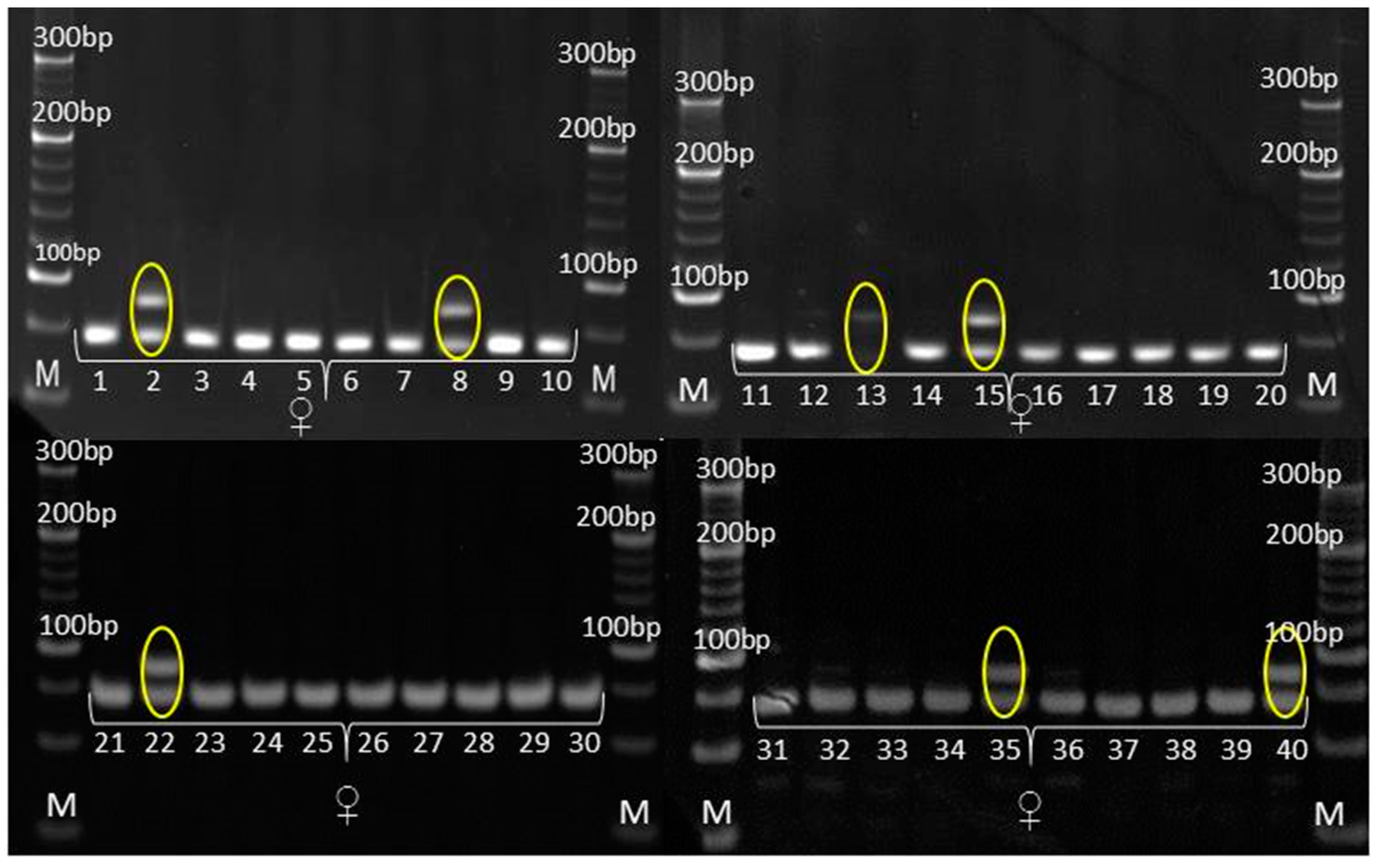
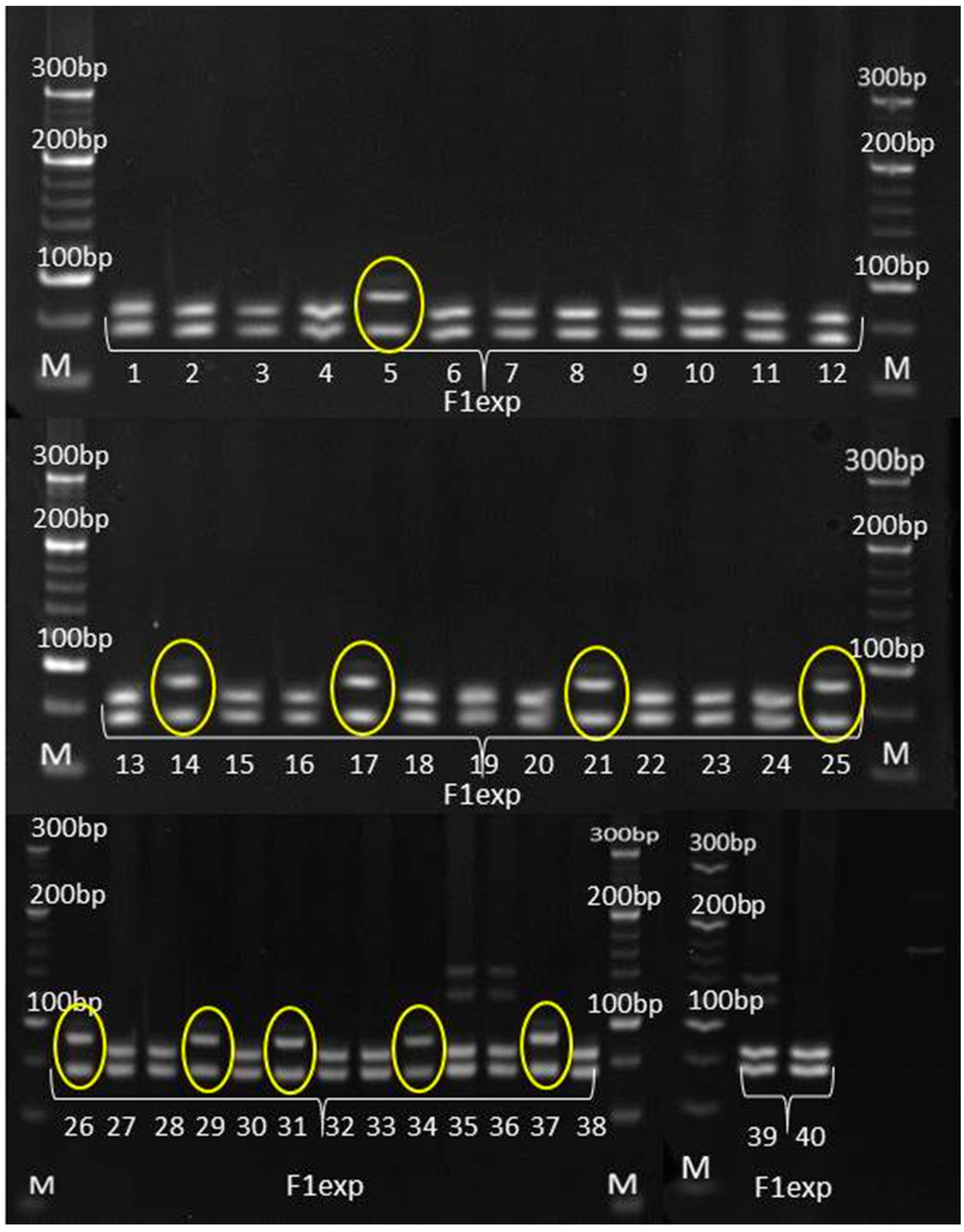
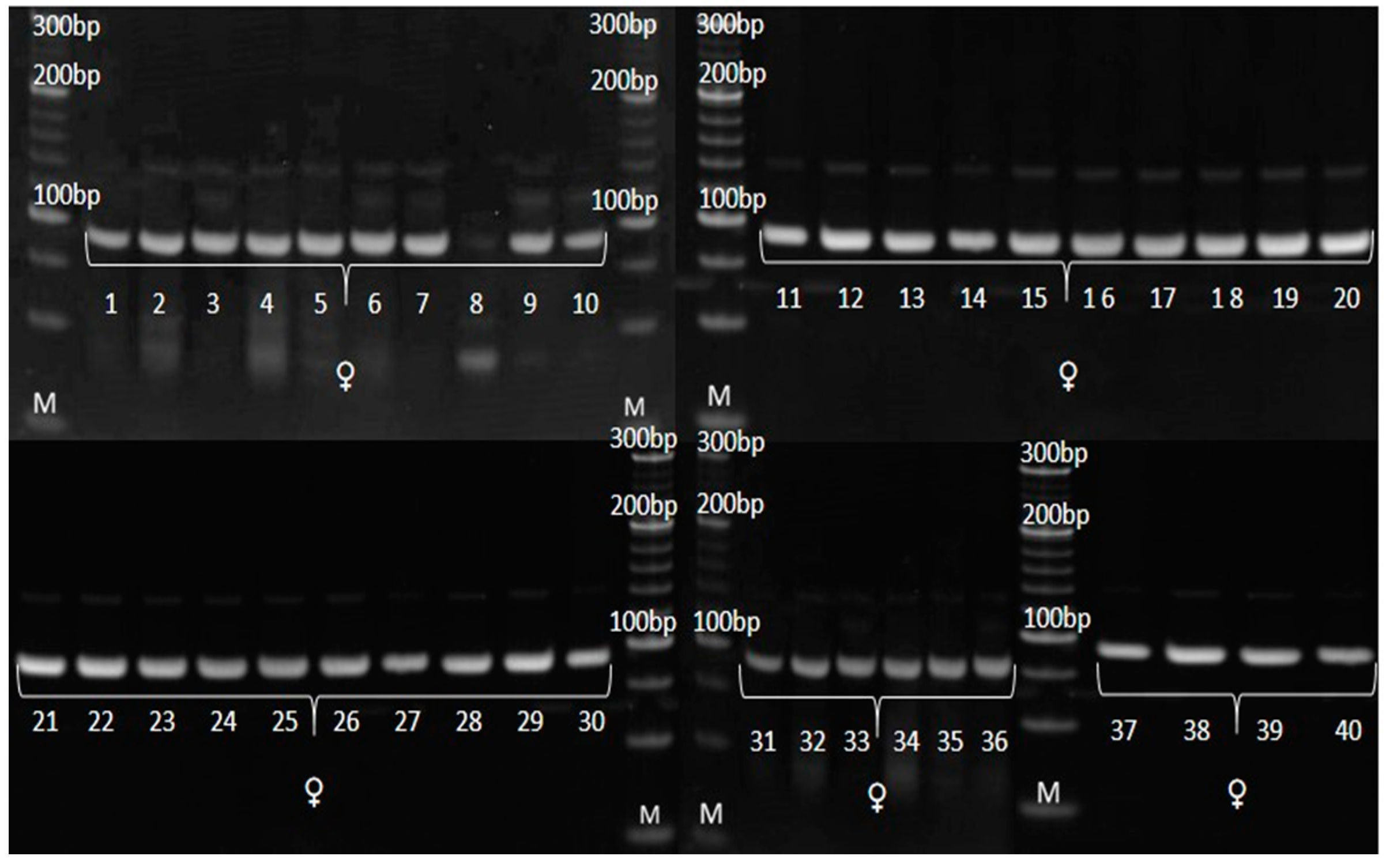
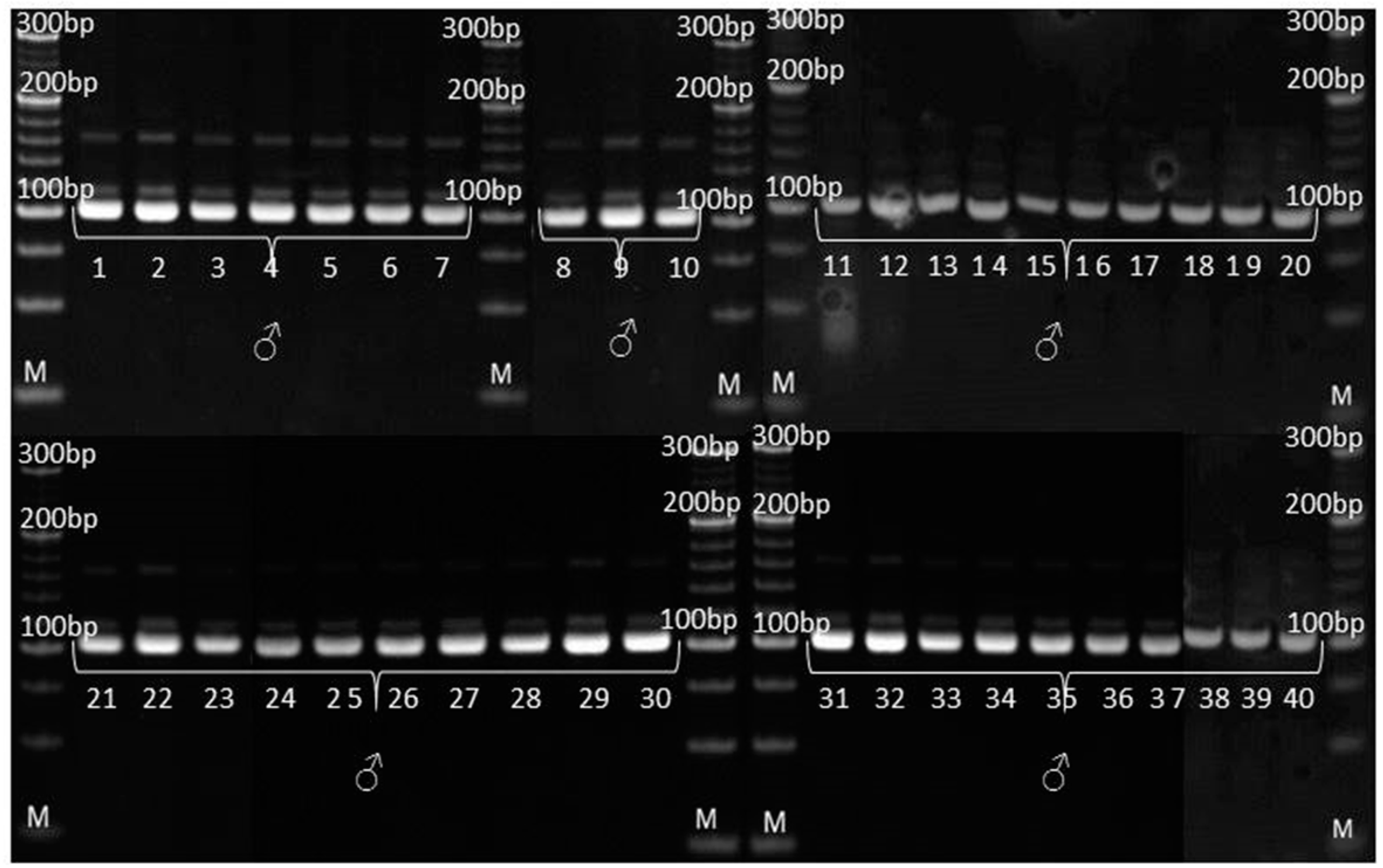
3.3. Genetic Purity Estimation by Molecular Markers
4. Discussion
4.1. Varietal Uniformity Estimation by Morphological Markers
4.2. Genetic Purity Estimation by Protein Markers
4.3. Genetic Purity Estimation by Molecular Markers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhat, M.I.; Gulzaffar; Dar, Z.A.; Lone, A.A.; Shikari, A.B.; Ali, G.; Wani, M.A.; Khan, G.H.; Gazal, A.; Lone, R.A. Genetic Purity Analysis in Maize under Temperate Conditions. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2710–2722. [Google Scholar] [CrossRef]
- Babić, V.; Marković, K.; Kravić, N.; Babić, M.; Srdić, J.; Petrović, T. Assessment of one maize hybrid lot uniformity by UPOV morphological and protein markers. Plant Breed. Seed Prod. 2022, 28, 23–33. [Google Scholar] [CrossRef]
- Song, L.; Liu, X.; Chen, W.; Hao, Z.; Bai, L.; Zhang, D. Genetic Relationships Among Chinese Maize OPVs Based on SSR Markers. J. Integr. Agric. 2013, 12, 1130–1137. [Google Scholar] [CrossRef]
- Law, J.R.; Anderson, S.R.; Jones, E.S.; Nelson, B.K.; Mulaosmanovic, E.; Smith, J.S. Characterization of maize germplasm: Comparison of morphological dataset compiled using different approaches to data recording. Maydica 2011, 56, 1710–1722. [Google Scholar]
- Daniel, I.O.; Adetumbi, J.A.; Oyelakin, O.O.; Olakojo, S.A.; Ajala, M.O.; Onagbesan, S.O. Application of SSR markers for genetic purity analysis of parental inbred lines and some commercial hybrid maize (Zea mays L.). Am. J. Exp. Agric. 2012, 2, 597–606. [Google Scholar] [CrossRef]
- Iqbal, J.; Shinwari, Z.K.; Rabbani, M.A. Investigation of total seed storage proteins of Pakistani and Japanese maize (Zea Mays L.) through SDS-PAGE markers. Pak. J. Bot. 2014, 46, 817–822. [Google Scholar]
- Ortiz, R.; Crossa, J.; Franco, J.; Sevilla, R.; Burgueño, J. Classification of Peruvian highland maize races using plant traits. Genet. Resour. Crop. Evol. 2008, 55, 151–162. [Google Scholar] [CrossRef]
- Dou, X.Y.; Yan, M.; Xu, Y.F.; Hussain, K.; Liu, Y.Z.; Lin, F. Identification of seed purity of maize hybrids by ultrathin layer isoelectric focusing electrophoresis of seed salt soluble proteins. Turk. J. Agric. For. 2012, 36, 267–273. [Google Scholar] [CrossRef]
- International Seed Testing Association (ISTA). Chapter 8: Species and variety testing (8.8.5). In International Rules for Seed Testing, Edition 2007; The International Seed Testing Association: Bassersdorf, Switzerland, 2007; Volume Full issue i-17, pp. 8: 18–29, ISBN-10 3-906549-38-0; ISBN-13 978-3-906549-38-5. [Google Scholar]
- Reddy, D.L.; Khandagale, K.; Victorathisayam, T.; Aswath, C.; Singh, T.H. Genetic purity of interspecific hybrids of Solanum melongena and S. macrocarpon assessed using SSR markers. Int. J. Veg. Sci. 2015, 21, 111–117. [Google Scholar] [CrossRef]
- UPOV. TG/2/7. Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability. Available online: https://www.upov.int/edocs/tgdocs/en/tg002.pdf (accessed on 10 February 2020).
- UPOV. TGP/10/2. Examining Uniformity. Available online: https://www.upov.int/edocs/tgpdocs/en/tgp_10.pdf (accessed on 8 May 2020).
- UPOV. TGP/8. Trial Design and Techniques Used in the Examination of Distinctness, Uniformity and Stability. Available online: https://www.upov.int/edocs/tgpdocs/en/tgp_8.pdf (accessed on 6 May 2020).
- International Seed Testing Association. Chapter 8: Species and variety testing (8.9.3). In International Rules for Seed Testing; Sutcliffe, V., Ed.; International Seed Testing Association: Bassersdorf, Switzerlanand, 2018; Full issue i-19-8; pp. 8: 9–13. Available online: https://www.heartwoodeditorial.co.uk. [CrossRef]
- International Seed Testing Association. Chapter 8: Species and variety testing (8.10.3). In International Rules for Seed Testing; Sutcliffe, V., Ed.; International Seed Testing Association: Bassersdorf, Switzerlanand, 2018; Full issue i-19-8; pp. 8: 24–25. Available online: https://www.heartwoodeditorial.co.uk. [CrossRef]
- Semagn, K.; Beyene, Y.; Makumbi, D.; Mugo, S.; Prasanna, B.M.; Magorokosho, C.; Altin, G. Quality control genotyping for assessment of genetic identity and purity in diverse tropical maize inbred lines. Theor. Appl. Genet. 2012, 125, 1487–1501. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Babić, V.; Nikolic, A.; Andjelkovic, V.; Kovacevic, D.; Filipovic, M.; Vasic, V.; Mladenovic-Drinic, S. UPOV morphological versus molecular markers for maize inbred lines variability determination. Chil. J. Agric. Res. 2016, 76, 417–426. [Google Scholar] [CrossRef]
- Yu, J.K.; Chung, Y.S. Plant Variety Protection: Current Practices and Insights. Genes 2021, 12, 1127. [Google Scholar] [CrossRef] [PubMed]
- Pham, B.; McConnaughay, K. Chapter 5: Plant Phenotypic Expression in Variable Environments. In Ecology and the Environment, The Plant Sciences, 3rd ed.; Monson, R.K., Ed.; Springer: New York, NY, USA, 2014; Volume 8, pp. 119–141. [Google Scholar] [CrossRef]
- Khairunnisa, H.; Puziak, H.; Shuhaimi, M. Corn silk (Stigma maydis) in healthcare: A phytochemical and pharmacological review. Molecules 2012, 17, 9697–9715. [Google Scholar] [CrossRef]
- Josia, C.; Mashingaidze, K.; Amelework, A.B.; Kondwakwenda, A.; Musvosvi, C.; Sibiya, J. SNP-based assessment of genetic purity and diversity in maize hybrid breeding. PLoS ONE 2021, 16, e0249505. [Google Scholar] [CrossRef]
- UPOV. General Introduction to the Examination of Distinctness, Uniformity and Stability and the Development of Harmonized Descriptions of New Varieties of Plants. Available online: https://www.upov.int/export/sites/upov/resource/en/tg_1_3.pdf (accessed on 18 April 2020).
- Kahriman, F.; Şerment, M.; Haşlak, M.; Kang, M.S. Pollen effect (xenia) for evaluating breeding materials in maize. Genet. Belgrade 2017, 49, 217–234. [Google Scholar] [CrossRef]
- Sinha, K.N.; Singh, M.; Kumar, C. Electrophoretic study of seed storage protein in five species of Bauhinia sp. Int. J. Pharma Biol. Sci. 2012, 4, 8–11. [Google Scholar] [CrossRef]
- Alireza, S.; Mohammad, F. Comparison of sunflower (Helianthus annuus L.) hybrids by using isozymes and seed storage protein. Biotechnology 2014, 13, 112–115. [Google Scholar] [CrossRef]
- Jiamtae, P.; Onwimol, D.; Chanprasert, W.; Thongket, T.; Prasertsak, A. Varietal verification of Thai rice using ultrathin layer isoelectric focusing of seed proteins. ScienceAsia 2017, 43, 145–154. [Google Scholar] [CrossRef]
- Kumar, M.B.A.; Sherry, R.J.; Varier, A.; Dadlani, M. Suitability of seed esterases for establishing distinctness, uniformity and stability of pearl millet genotypes. Curr. Sci. 2007, 93, 951–956. [Google Scholar]
- Van den Berg, B.M. Inbred testing of tomato (Lycopersicon esculentum L.) F1 varieties by ultrathin-layer isoelectric focusing of seed protein. Electrophoresis 1990, 11, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Yan, M. Genetic purity testing of pumpkin hybrid seed by ultrathin-layer isoelectric focusing. Crop. Breed. Appl. Biotechnol. 2013, 13, 200–202. [Google Scholar] [CrossRef]
- Lucchese, C.; Dinelli, G.; Miggiano, A.; Lovato, A. Identification of pepper (Capsicum spp.) cultivars by field and electrophoresis tests. Seed Sci. Technol. 1999, 27, 37–47. [Google Scholar]
- Wang, X.F.; Knoblauch, R.; Leist, N. Identification of varieties and testing of hybrid purity of rice by ultrathin-layer isoelectric focusing of seed protein. Int. Rice Res. Notes 2001, 26, 18–19. [Google Scholar]
- Zhao, T.; Yan, M.; Lu, Y.P.; Yang, F.; Huang, J.; Wang, X.F. Genetic purity testing of two-line hybrid rice seeds by ultrathin-layer isoelectric focusing of proteins. Seed Sci. Technol. 2005, 33, 45–52. [Google Scholar] [CrossRef]
- Yan, M.; Ye, H.; Wang, X.F. Investigation into the feasibility of assessing the hybridity of immature rice grains. Seed Sci. Technol. 2006, 34, 1–8. [Google Scholar] [CrossRef]
- Smith, J.S.C.; Register, J.C. Genetic purity and testing technologies for seed quality: A company perspective. Seed Sci. Res. 1998, 8, 285–294. [Google Scholar] [CrossRef]
- Gethi, J.G.; Labate, J.A.; Lamkey, K.R.; Smith, M.E.; Kresovich, S. SSR variation in important U.S. maize inbred lines. Crop. Sci. 2002, 42, 951–957. [Google Scholar] [CrossRef]
- Hamblin, M.T.; Warburton, M.L.; Buckler, E.S. Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS ONE 2007, 2, e1367. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Ramekar, R.V.; Sa, K.J.; Woo, S.Y.; Lee, J.K. Non-parental banding patterns in recombinant inbred line population of maize with SSR markers. Genet. Mol. Res. 2015, 14, 8420–8430. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic distribution, putative functions, and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, Y.; Jaqueth, J.S.; Matsuoka, Y.; Smith, O.S.; Beavis, W.D.; Smith, J.S.C.; Doebley, J. Rate and pattern of mutation at microsatellite loci in maize. Mol. Biol. Evol. 2002, 19, 1251–1260. [Google Scholar] [CrossRef]
- Vigouroux, Y.; Mitchell, S.; Matsuoka, Y.; Hamblin, M.; Kresovich, S.; Smith, J.S.C.; Jaqueth, J.; Smith, O.S.; Doebley, J. An analysis of genetic diversity across the maize genome using microsatellites. Genetics 2005, 169, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Raskina, O.; Barber, J.C.; Nevo, E.; Belyayev, A. Repetitive DNA and chromosomal rearrangements: Speciation related events in plant genomes. Cytogenet. Genome Res. 2008, 120, 351–357. [Google Scholar] [CrossRef]
- Hua-Van, A.; Le Rouzic, A.; Boutin, T.S.; Filée, J.; Capy, P. The struggle for life of the genome’s selfish architects. Biol. Direct 2011, 6, 19. [Google Scholar] [CrossRef]
- Chadha, S.; Sharma, M. Transposable elements as stress adaptive capacitors induce genomic instability in fungal pathogen Magnaporthe oryzae. PLoS ONE 2014, 9, e94415. [Google Scholar] [CrossRef]
- Lee, J.K.; Park, J.Y.; Kim, J.H.; Kwon, S.J.; Shin, J.H.; Hong, S.K.; Min, H.K.; Kim, N.S. Genetic mapping of the Issac-CACTA transposon in maize. Theor. Appl. Genet. 2006, 113, 16–22. [Google Scholar] [CrossRef]
- Li, L.; Liu, L.; Zhang, D.S.; Wu, P.; Zhang, F.L.; Xu, X.L. Hybrid purity testing of Brassica rapa using SSR marker technology. Hortic. Sci. 2017, 52, 1342–1348. [Google Scholar] [CrossRef]
- Yashitola, J.; Thirumurugan, T.; Sundaram, R.M.; Naseerullah, M.K.; Ramesha, M.S.; Sarma, N.P.; Sonti, R.V. Assessment of purity of rice hybrids using microsatellite and STS markers. Crop. Sci. 2002, 42, 1369–1373. [Google Scholar] [CrossRef]
- Nandakumar, N.; Singh, A.K.; Sharma, R.K.; Mohapatra, T.; Prabhu, K.V.; Zaman, F.U. Molecular fingerprinting of hybrids and assessment of genetic purity of hybrid seeds in rice using microsatellite markers. Euphytica 2004, 136, 257–264. [Google Scholar] [CrossRef]
- Warburton, M.L.; Setimela, P.; Franco, J.; Cordova, H.; Pixley, K.; Bänziger, M.; Dreisigacker, S.; Bedoya, C.; Macrobert, J. Toward a cost-effective fingerprinting methodology to distinguish maize open-pollinated varieties. Crop. Sci. 2010, 50, 467–477. [Google Scholar] [CrossRef]
- Ertiro, B.T.; Semagn, K.; Das, B.; Olsen, M.; Labuschagne, M.; Worku, M. Genetic variation and population structure of maize inbred lines adapted to the mid-altitude sub-humid maize agro-ecology of Ethiopia using single nucleotide polymorphic (SNP) markers. BMC Genom. 2017, 18, 777–779. [Google Scholar] [CrossRef] [PubMed]

| No | List of Morphological Traits | DSA | TO | RND |
|---|---|---|---|---|
| 3 | Intensity of leaf green color | Inflorescence visible | VS | 1–3 |
| 4 | Undulation of leaf blade margin | Inflorescence visible | VS | 1–3 |
| 5 | Angle between blade and stem | Anthesis | VS | 1–9 |
| 6 | Curvature of leaf blade | Anthesis | VS | 1–9 |
| 7 | Degree of stem zig-zag | Anthesis | VS | 1–3 |
| 9 | Anth. col. at base of tassel glume | Anthesis | VS | 1–9 |
| 10 | Anth. col. of tassel glume exclude base | Anthesis | VS | 1–9 |
| 11 | Anth. col. of tassel anthers | Anthesis | VS | 1–9 |
| 12 | Angle btw. main axis and lateral tass. br. | Anthesis | VS | 1–9 |
| 13 | Curvature of lateral tass. branches | Anthesis | VS | 1–9 |
| 14 | Number of primary tassel branches | Anthesis to milk devel. | MS | no. |
| 16 | Anthocyanin coloration of silks | Anthesishafway | VS | 1–9 |
| 17 | Anthocyanin coloration of brace roots | Anthesis to milk devel. | VS | 1–9 |
| 18 | Density of tassel spikelets | Anthesis to watery ripe | VS | 3–7 |
| 19 | Anthocyanin coloration of sheath | Watery ripe to milk | VS | 1–9 |
| 20 | Anthocyanin coloration of internodes | Watery ripe to milk | VS | 1–9 |
| 21 | Length of main t. axis above lowest l. b. | Watery ripe to milk | MS | cm |
| 22 | Length of main t. axis above upper l. b. | Watery ripe to milk | MS | cm |
| 23 | Length of lateral branch | Watery ripe to milk | VS | 1–9 |
| 24 | Height of plant | Milk to dough develop. | MS | cm |
| 25 | Ratio Height of ear/ Height of plant | Milk to dough develop. | MS | cm |
| 26 | Width of blade | Milk to dough develop. | MS | cm |
| 27 | Length of ear peduncle | Milk to dough develop. | VS | 1–9 |
| 28 | Length of ear | After harvest | MS | cm |
| 29 | Diameter of ear in middle | After harvest | MS | cm |
| 30 | Shape of ear | After harvest | VS | 1–3 |
| 31 | Number of rows of grain | After harvest | MS | no |
| 36 | Type of grain | After harvest | VS | 1–9 |
| 38 | Color of top of grain | After harvest | VS | 1–9 |
| 39 | Color of dorsal side of grain | After harvest | VS | 1–9 |
| 41 | Anthocyanin coloration of glumes of cob | After harvest | VS | 1–9 |
| SSR Markers | Bin | Repeat | Forward Sequence (5’-3’) | Reverse Sequence (5’-3’) |
|---|---|---|---|---|
| phi109275 | 1.03 | AGCT | CGGTTCATGCTAGCTCTGC | GTTGTGGCTGTGGTGGTG |
| phi083 | 2.04 | AGCT | CAAACATCAGCCAGAGACAAGGAC | ATTCATCGACGCGTCACAGTCTACT |
| umc1448 | 2.04 | (GCT)5 | ATCCTCTCATCTTTAGGTCCACCG | CATATACAGTCTCTTCTGGCTGCTCA |
| phi102228 | 3.06 | AAGC | ATTCCGACGCAATCAACA | TTCATCTCCTCCAGGAGCCTT |
| umc1117 | 4.04 | (TCGCA)4 | AATTCTAGTCCTGGGTCGGAACTC | CGTGGCCGTGGAGTCTACTACT |
| umc1478 | 5.01 | (GGAG)4 | GAAGCTTCTCCTCTCGCGTCTC | CAGTCCCAGACCCTAGCTCAGTC |
| umc1133 | 6.01 | ATAC | ATTCGATCTAGGGTTTGGGTTCAG | GATGCAGTAGCATGCTGGATGTAG |
| umc1545 | 7.00 | (AAGA)4 | GAAAACTGCATCAACAACAAGCTG | ATTGGTTGGTTCTTGCTTCCATTA |
| phi015 | 8.08 | AAAC | GCAACGTACCGTACCTTTCCGA | ACGCTGCATTCAATTACCGGGAAG |
| umc1061 | 10.06 | (TCG)6 | AGCAGGAGTACCCATGAAAGTCC | TATCACAGCACGAAGCGATAGATG |
| No | Plant Trait | Mean Value | STDEV Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F1c | F1exp | Lc | F1♀ | F1♂ | F1c | F1exp | Lc | F1♀ | F1♂ | ||
| 14 | NPTB | 7.8 | 7.2 | 7.4 | 6.8 | 1.42 | 2.7 | 1.7 | 1.3 | 1.41 | 0.8 |
| 21 | PHLTLB | 206.2 | 190.7 | 161.6 | 175.7 | 92.0 | 18.9 | 18.5 | 13.8 | 10.1 | 13.0 |
| 22 | PHHTLB | 217.1 | 203.3 | 175.2 | 183.6 | 92.3 | 17.8 | 17.7 | 14.1 | 10.4 | 13.2 |
| 24 | HP | 242.1 | 229.0 | 196.5 | 206.3 | 114.9 | 18.1 | 17.8 | 14.3 | 11.2 | 13.6 |
| 25 | HE | 104.5 | 63.1 | 70.5 | 58.2 | 27.4 | 10.2 | 9.3 | 9.0 | 8.1 | 4.7 |
| 26 | WB | 9.6 | 9.7 | 9.15 | 7.4 | 6.6 | 1.6 | 1.7 | 0.8 | 0.5 | 0.7 |
| 28 | LE | 19.5 | 18.8 | 18.1 | 13.3 | 8.6 | 1.7 | 1.9 | 2.4 | 2.0 | 1.8 |
| 29 | DE | 4.5 | 4.2 | 3.3 | 3.6 | 2.9 | 0.3 | 0.2 | 0.3 | 0.3 | 0.2 |
| 31 | NR | 14.4 | 14.7 | 14.2 | 13.5 | 10.9 | 1.2 | 1.4 | 1.7 | 1.5 | 1.6 |
| No | List of Morphological Traits | Note | Off-Type Note | ||||
|---|---|---|---|---|---|---|---|
| F1exp | F1♀ | F1♂ | F1exp | F1♀ | F1♂ | ||
| 3 | ILGC | 2 | 2 | 1 | |||
| 4 | ULBM | 1 | 1 | 1 | |||
| 5 | ABLB | 1 | 1 | 3 | |||
| 6 | CLB | 3 | 1 | 1 | |||
| 7 | DSZ | 1 | 1 | 1 | |||
| 9 | ACBG | 1 | 1 | 1 | |||
| 10 | ACG | 3 | 3 | 1 | – | – | 3-1p |
| 11 | ACA | 5 | 5 | 1 | 1-1p; 7-1p | 1-2p | – |
| 12 | ABMALB | 5 | 3 | 1 | |||
| 13 | CLTB | 3 | 5 | 1 | |||
| 16 | ACS | 5-16p; 3-20p | 1-17p; 3-23p | 3 | 1-3p; 7-1p | – | – |
| 17 | ACBR | 5-31p; 7-8p | 5-28p; 7-10p | 7(0*) | 3-1p | 3-2p | – |
| 18 | DTS | 3 | 5 | 1 | – | 3-1p | – |
| 19 | ACSh | 1 | 1 | 1 | |||
| 20 | ACIn | 1 | 1 | 3-17p; 5-23p | |||
| 23 | LLB | 5 | 5 | 3 | 9-1p | – | – |
| 27 | LEP | 5 | 5 | 1 | |||
| 30 | ShE | 2 | 2 | 3 | |||
| 36 | TG | 4-13p; 5-22p | 5 | 1 | 3-5p | – | – |
| 38 | CTG | 3-26p;4-14p | 3 | 4 | – | 4-4p | – |
| 39 | CDSG | 5 | 5 | 4 | 4-1p | – | – |
| 41 | ACGCb | 1 | 1 | 1 | |||
| Σ Off type plants | 14p | 9p | 1p | ||||
| Primer | F1♂ | F1♀ | F1exp |
|---|---|---|---|
| phi109275 | * | * | * |
| phi083 | * | * | * |
| umc1448 | * | * | * |
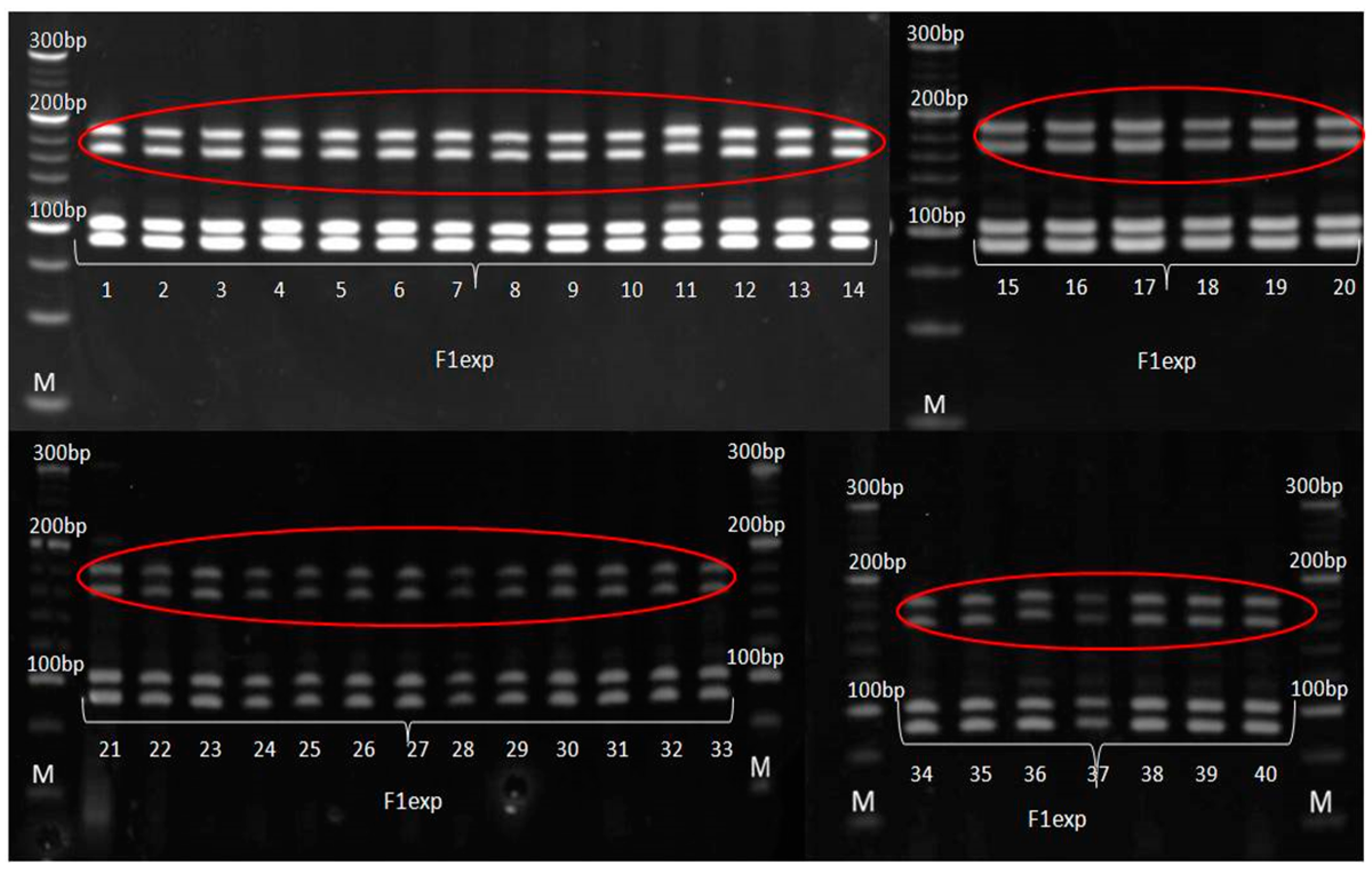
| phi102228 | * | * | non-parental bands in all |
| umc1117 | * | * | non-parental bands |
| umc1478 | * | * | non-parental bands |
| umc1133 | uniform | uniform | non-parental bands in all |
| umc1545 | uniform | non-specific band | F1♀ non-specific band |
| phi015 | uniform | uniform | non-parental band in all |
| umc1061 | * | * | non-parental bands |
| Primer | F1♂ | F1♀ | F1exp | F1exp Non-Specific/Non-Parental Band | F1♀ Non-Specific Band |
|---|---|---|---|---|---|
| phi109275 | 130 bp | 130 bp | 130 bp | – | – |
| phi083 | 202 bp | 202 bp | 202 bp | – | – |
| umc1448 | 154 bp | 154 bp | 154 bp | – | – |
| phi102228 | 138 bp | 138 bp | 138 bp | 155/185 bp | – |
| umc1117 | 120/125 bp | 120/125 bp | 120/125 bp | 202/223 bp | – |
| umc1478 | 135/140 bp | 135 bp | 135 bp | 195/245 bp | –– |
| umc1133 | 102 bp | 91 bp | 91/102 bp | 165/184 bp | – |
| umc1545 | 85 bp | 77 bp | 77/85 bp | 92 bp | 92 bp |
| phi015 | 135 bp | 127 bp | 127/135 bp | 187 bp | – |
| umc 1061 | 106 bp | 106 bp | 106 bp | 125/137 bp | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovincic, A.; Markovic, K.; Ristic, D.; Babic, V.; Petrovic, T.; Zivanovic, T.; Kravic, N. Efficiency of Biological Typing Methods in Maize Hybrid Genetic Purity Estimation. Genes 2023, 14, 1195. https://doi.org/10.3390/genes14061195
Kovincic A, Markovic K, Ristic D, Babic V, Petrovic T, Zivanovic T, Kravic N. Efficiency of Biological Typing Methods in Maize Hybrid Genetic Purity Estimation. Genes. 2023; 14(6):1195. https://doi.org/10.3390/genes14061195
Chicago/Turabian StyleKovincic, Anika, Ksenija Markovic, Danijela Ristic, Vojka Babic, Tanja Petrovic, Tomislav Zivanovic, and Natalija Kravic. 2023. "Efficiency of Biological Typing Methods in Maize Hybrid Genetic Purity Estimation" Genes 14, no. 6: 1195. https://doi.org/10.3390/genes14061195
APA StyleKovincic, A., Markovic, K., Ristic, D., Babic, V., Petrovic, T., Zivanovic, T., & Kravic, N. (2023). Efficiency of Biological Typing Methods in Maize Hybrid Genetic Purity Estimation. Genes, 14(6), 1195. https://doi.org/10.3390/genes14061195





