Analysis of the Origin of Emiratis as Inferred from a Family Study Based on HLA-A, -C, -B, -DRB1, and -DQB1 Genes
Abstract
1. Introduction
2. Material and Methods
2.1. Study Subjects
2.2. DNA Genotyping
2.3. Statistical Analysis
3. Results
3.1. HLA Allele Lineage Frequencies
3.2. Two- and Five-Locus HLA Haplotypes
3.3. Ewens–Watterson Homozygosity Test of Neutrality
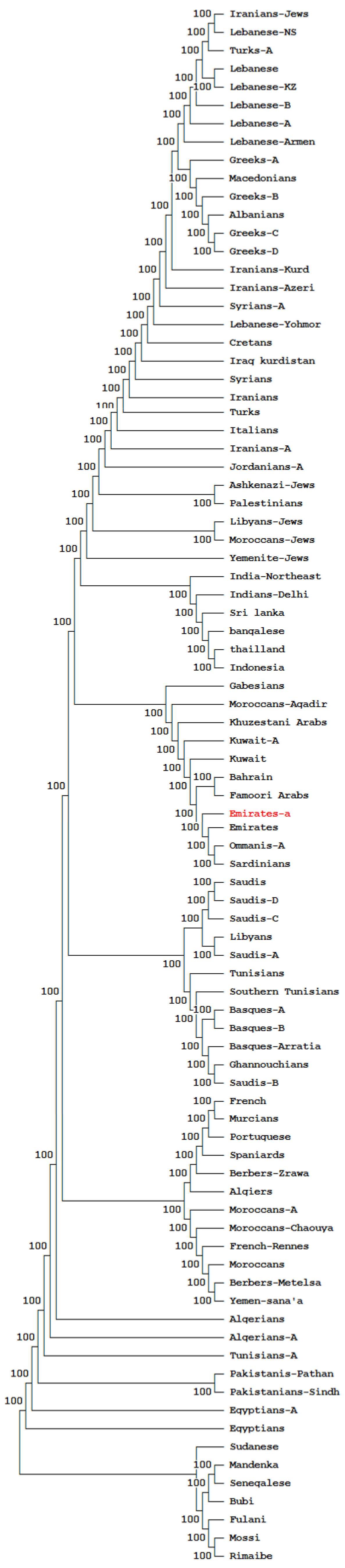
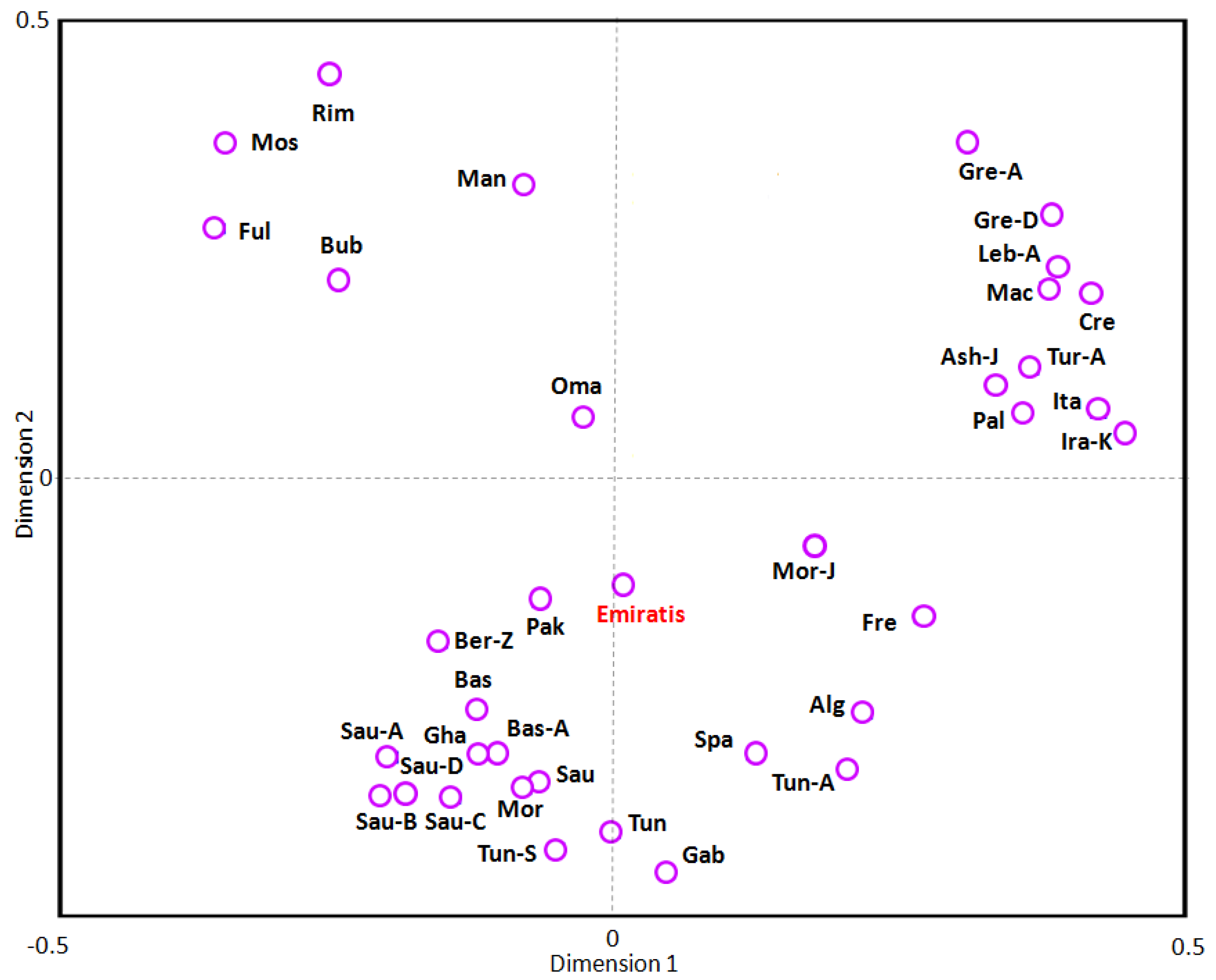
3.4. Phylogenetic and Correspondence Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- HLA Database. 2019. Available online: http://hla.alleles.org (accessed on 20 March 2020).
- Marrack, P.; Kappler, J. The antigen-specific, major histocompatibility complex-restricted receptor on T cells. Adv. Immunol. 1986, 38, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Phylogenetic analysis in molecular evolutionary genetics. Annu. Rev. Genet. 1996, 30, 371–403. [Google Scholar] [CrossRef]
- Gourraud, P.A.; Barnetche, T.; Vidan-Jeras, B.; Cambon-Thomsen, A. Introduction to statistical analysis of population data in immunogenetics. Transpl. Immunol. 2005, 14, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Worldometers Counter. 2020. Available online: https://www.worldometers.info/world-population/united-arab-emirates-population/ (accessed on 2 April 2020).
- United Arab Emirates Population Statistics 2023. Available online: https://www.globalmediainsight.com/blog/uae-population-statistics/ (accessed on 1 April 2023).
- Wikipedia Encyclopedia. Available online: https://en.wikipedia.org/wiki/United_Arab_Emirates (accessed on 27 July 2021).
- Armitage, S.J.; Jasim, S.A.; Marks, A.E.; Parker, A.G.; Usik, V.I.; Uerpmann, H.P. The southern route “out of Africa”: Evidence for an early expansion of modern humans into Arabia. Science 2011, 331, 453–456. [Google Scholar] [CrossRef]
- Quintana-Murci, L.; Semino, O.; Bandelt, H.J.; Passarino, G.; McElreavey, K.; Santachiara-Benerecetti, A.S. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat. Genet. 1999, 23, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Hajjej, A.; Almawi, W.Y.; Hattab, L.; El-Gaaied, A.; Hmida, S. The investigation of the origin of Southern Tunisians using HLA genes. J. Hum. Genet. 2017, 62, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W. Gametic Disequilibrium Measures: Proceed With Caution. Genetics 1987, 117, 331–341. [Google Scholar] [CrossRef]
- Cramer, H. Mathematical Methods of Statistics; Princeton University Press: Princeton, NJ, USA, 1946. [Google Scholar]
- Single, R.M.; Strayer, N.; Thomson, G.; Paunic, V.; Albrecht, M.; Maiers, M. Asymmetric Linkage Disequilibrium: Tools for Assessing Multiallelic LD. Hum. Immunol. 2016, 77, 288–294. [Google Scholar] [CrossRef]
- Osoegawa, K.; Mack, S.J.; Prestegaard, M.; Fernández-Viña, M.A. Tools for Building, Analyzing and Evaluating HLA Haplotypes from Families. Hum. Immunol. 2019, 80, 633–643. [Google Scholar] [CrossRef]
- Osoegawa, K.; Mallempati, K.C.; Gangavarapu, S.; Oki, A.; Gendzekhadze, K.; Marino, S.R.; Brown, N.K.; Bettinotti, M.P.; Weimer, E.T.; Montero-Martín, G.; et al. HLA alleles and haplotypes observed in 263 US families. Hum. Immunol. 2019, 80, 644–660. [Google Scholar] [CrossRef]
- Lancaster, A.K.; Nelson, M.P.; Single, R.M.; Meyer, D.; Thomson, G. PyPop: A Software Framework for Population Genomics: Analyzing Large-Scale Multi-Locus Genotype Data. In Pacific Symposium on Biocomputing 8; Altman, R.B., Dunker, K., Hunter, L., Jung, T., Klein, T., Eds.; World Scientific: Singapore, 2003; pp. 514–525. [Google Scholar]
- Lancaster, A.K.; Single, R.M.; Solberg, O.D.; Nelson, M.P.; Thomson, G. PyPop update—A software pipeline for large-scale multilocus population genomics. Tissue Antigens 2007, 69, 192–197. [Google Scholar] [CrossRef]
- Ewens, W. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 1972, 3, 87–112. [Google Scholar] [CrossRef]
- Watterson, G. The homozygosity test of neutrality. Genetics 1978, 88, 405–417. [Google Scholar] [CrossRef]
- Slatkin, M. An exact test for neutrality based on the Ewens sampling distribution. Genet. Res. 1994, 64, 71–74. [Google Scholar] [CrossRef]
- Slatkin, M. A correction to the exact test based on the Ewens sampling distribution. Genet. Res. 1996, 68, 259–260. [Google Scholar] [CrossRef]
- Nei, M.; Tajima, Y.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gen frequency data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy; Freeman: San Francisco, CA, USA, 1973. [Google Scholar]
- Nei, M. Genetic distances between populations. Am. Nat. 1972, 106, 283. [Google Scholar] [CrossRef]
- Young, F.W.; Bann, C.M. A Visual Statistics System. In Statistical Computing Environments for Social Researches; Stine, R.A., Fox, J., Eds.; Sage Publications: New York, NY, USA, 1996; pp. 207–236. [Google Scholar]
- Solberg, O.D.; Mack, S.J.; Lancaster, A.K.; Single, R.M.; Tsai, Y.; Sanchez-Mazas, A.; Thomson, G. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: A meta-analytic review of 497 population studies. Hum. Immunol. 2008, 69, 443–464. [Google Scholar] [CrossRef]
- Valluri, V.; Mustafa, M.; Santhosh, A.; Middleton, D.; Alvares, M.; El Haj, E.; Gumama, O.; Abdel-Wareth, L. Frequencies of HLA-A, HLA-B, HLA-DR, and HLA-DQ phenotypes in the United Arab Emirates population. Tissue Antigens 2005, 66, 107–113. [Google Scholar] [CrossRef]
- Kulski, J.K.; AlSafar, H.S.; Mawart, A.; Henschel, A.; Tay, G. HLA class I allele lineages and hapltype frequencies in Arabs of the United Arab Emirates. Int. J. Immunogenet. 2019, 46, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz-Villena, A.; Yafei, Z.A.; Juarez, I.; Palacio-Gruber, J.; Mahri, A.A.; Alvares, M.; Lopez-Nares, A.; Nieto, J.; Al Seiari, M.; Martin-Villa, J.M.; et al. HLA genetic study from United Arab Emirates (UAE), Abu Dhabi. Hum. Immunol. 2019, 80, 421–422. [Google Scholar] [CrossRef] [PubMed]
- Alnaqbi, H.; Tay, G.; Chehadeh, S.; Al Safar, H. Characterizing the diversity of MHC conserved extended haplotypes using families from the United Arab Emirates. Sci. Rep. 2022, 12, 7165. [Google Scholar] [CrossRef] [PubMed]
- Database of Allele Frequencies. 2019. Available online: http://www.allelefrequencies.net (accessed on 30 March 2020).
- Wallace, G.R. HLA-B* 51 the primary risk in Behçet disease. Proc. Natl. Acad. Sci. USA 2014, 111, 8706–8707. [Google Scholar] [CrossRef] [PubMed]
- Al Yafei, Z.; Mack, S.J.; Alvares, M.; Ali, B.R.; Al Kaabi, J.; ElGhazali, G. HLA-DRB1 and –DQB1 alleles, haplotypes and genotypes in Emirati patients with Type 1 Diabetes. Front. Genet. 2022, 13, 841879. [Google Scholar] [CrossRef] [PubMed]
- Ameen, R.; Shemmari, S.A.; Askar, M. Next-generation sequencing characterization of HLA in multi-generation families of Kuwaiti descent. Hum. Immunol. 2018, 79, 137–142. [Google Scholar] [CrossRef]
- Ameen, R.; Al Shemmari, S.H.; Marsh, S.G.E. HLA haplotype frequencies and genetic profiles of the kuwaiti population. Med. Princ. Pract. 2020, 29, 39–45. [Google Scholar] [CrossRef]
- NMDP/Be the Match. Available online: https://www.haplostats.org/haplostats?execution=e1s1 (accessed on 21 July 2022).
- Santhosh, A.; Mustafa, M.I.; Vijayalakshmi, V. HLA A-B and HLA A-B-DR haplotype frequencies in United Arab Emirates nationals. Emirates Med. J. 2004, 22, 215–220. [Google Scholar]
- Arnaiz-Villena, A.; Gomez-Casado, E.; Martinez-Laso, J. Population genetic relationships between Mediterranean populations determined by HLA allele distribution and a historic Perspective. Tissue Antigens 2002, 60, 111–121. [Google Scholar] [CrossRef]

| A-Locus | B-Locus | C-Locus | DRB1-Locus | DQB1-Locus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Frequency | SD * | Allele | Frequency | SD* | Allele | Frequency | SD * | Allele | Frequency | SD * | Allele | Frequency | SD * |
| A*02 | 0.2225 | 0.0208 | B*51 | 0.1950 | 0.0198 | C*07 | 0.2000 | 0.0200 | DRB1*03 | 0.2225 | 0.0208 | DQB1*02 | 0.3275 | 0.0235 |
| A*11 | 0.0950 | 0.0147 | B*35 | 0.1375 | 0.0172 | C*04 | 0.1550 | 0.0181 | DRB1*16 | 0.1775 | 0.0191 | DQB1*05 | 0.2975 | 0.0229 |
| A*26 | 0.0900 | 0.0143 | B*08 | 0.1150 | 0.0160 | C*06 | 0.1375 | 0.0172 | DRB1*07 | 0.1275 | 0.0167 | DQB1*03 | 0.2025 | 0.0201 |
| A*01 | 0.0850 | 0.0140 | B*50 | 0.0825 | 0.0138 | C*15 | 0.1300 | 0.0168 | DRB1*04 | 0.1025 | 0.0152 | DQB1*06 | 0.1400 | 0.0174 |
| A*03 | 0.0800 | 0.0136 | B*58 | 0.0575 | 0.0117 | C*12 | 0.0900 | 0.0143 | DRB1*15 | 0.0975 | 0.0149 | DQB1*04 | 0.0325 | 0.0089 |
| A*24 | 0.0775 | 0.0134 | B*40 | 0.0500 | 0.0109 | C*03 | 0.0675 | 0.0126 | DRB1*11 | 0.0700 | 0.0128 | |||
| A*32 | 0.0750 | 0.0132 | B*14 | 0.0450 | 0.0104 | C*16 | 0.0675 | 0.0126 | DRB1*13 | 0.0550 | 0.0114 | |||
| A*33 | 0.0675 | 0.0126 | B*52 | 0.0350 | 0.0092 | C*08 | 0.0450 | 0.0104 | DRB1*01 | 0.0450 | 0.0104 | |||
| A*68 | 0.0675 | 0.0126 | B*18 | 0.0325 | 0.0089 | C*14 | 0.0300 | 0.0085 | DRB1*10 | 0.0375 | 0.0095 | |||
| A*30 | 0.0550 | 0.0114 | B*15 | 0.0300 | 0.0085 | C*02 | 0.0300 | 0.0085 | DRB1*08 | 0.0225 | 0.0074 | |||
| A*31 | 0.0300 | 0.0085 | B*41 | 0.0250 | 0.0078 | C*17 | 0.0275 | 0.0082 | DRB1*14 | 0.0225 | 0.0074 | |||
| A*23 | 0.0250 | 0.0078 | B*53 | 0.0225 | 0.0074 | C*01 | 0.0125 | 0.0056 | DRB1*12 | 0.0125 | 0.0056 | |||
| A*74 | 0.0100 | 0.0050 | B*37 | 0.0200 | 0.0070 | C*05 | 0.0050 | 0.0035 | DRB1*09 | 0.0075 | 0.0043 | |||
| A*66 | 0.0075 | 0.0043 | B*73 | 0.0175 | 0.0066 | C*18 | 0.0025 | 0.0025 | ||||||
| A*34 | 0.0050 | 0.0035 | B*57 | 0.0175 | 0.0066 | |||||||||
| A*29 | 0.0050 | 0.0035 | B*07 | 0.0175 | 0.0066 | |||||||||
| A*69 | 0.0025 | 0.0025 | B*49 | 0.0175 | 0.0066 | |||||||||
| B*44 | 0.0150 | 0.0061 | ||||||||||||
| B*38 | 0.0125 | 0.0056 | ||||||||||||
| B*42 | 0.0125 | 0.0056 | ||||||||||||
| B*27 | 0.0100 | 0.0050 | ||||||||||||
| B*45 | 0.0100 | 0.0050 | ||||||||||||
| B*13 | 0.0075 | 0.0043 | ||||||||||||
| B*39 | 0.0050 | 0.0035 | ||||||||||||
| B*56 | 0.0025 | 0.0025 | ||||||||||||
| B*47 | 0.0025 | 0.0025 | ||||||||||||
| B*81 | 0.0025 | 0.0025 | ||||||||||||
| B*55 | 0.0025 | 0.0025 | ||||||||||||
| Loc1~Loc2 | D’ | Wn | WLoc1/Loc2 | WLoc2/Loc1 | N_Haplotypes |
|---|---|---|---|---|---|
| A~B | 0.480 | 0.373 | 0.323 | 0.397 | 140 |
| A~C | 0.414 | 0.312 | 0.319 | 0.303 | 112 |
| A~DRB1 | 0.309 | 0.235 | 0.239 | 0.234 | 117 |
| A~DQB1 | 0.208 | 0.224 | 0.220 | 0.128 | 64 |
| B~C | 0.845 | 0.749 | 0.783 | 0.637 | 65 |
| B~DRB1 | 0.5215 | 0.418 | 0.482 | 0.337 | 124 |
| B~DQB1 | 0.432 | 0.469 | 0.469 | 0.219 | 84 |
| C~DRB1 | 0.406 | 0.280 | 0.327 | 0.317 | 96 |
| C~DQB1 | 0.308 | 0.319 | 0.305 | 0.193 | 54 |
| DRB1~DQB1 | 0.907 | 0.820 | 0.895 | 0.583 | 26 |
| Haplotype | Frequency | SD * | D’ | χ2 | p-Value | Haplotype | Frequency | SD * | D’ | χ2 | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A~B | B~C | ||||||||||
| A*02~B*51 | 0.0750 | 0.0132 | 0.21 | 14.72 | 1.0 × 10−4 | B*41~C*17 | 0.0150 | 0.0061 | 0.59 | 125.70 | <1.0 × 10−5 |
| A*26~B*08 | 0.0525 | 0.0112 | 0.53 | 85.26 | 1.0 × 10−5 | B*53~C*04 | 0.0150 | 0.0061 | 0.61 | 18.40 | 1.8 × 10−5 |
| A*33~B*58 | 0.0250 | 0.0078 | 0.39 | 52.30 | <1.0 × 10−5 | B*07~C*07 | 0.0150 | 0.0061 | 0.82 | 19.23 | 1.2 × 10−5 |
| A*03~B*50 | 0.0250 | 0.0078 | 0.25 | 24.31 | <1.0 × 10−5 | B*57~C*06 | 0.0125 | 0.0056 | 0.67 | 19.99 | <1.0 × 10−5 |
| A*11~B*40 | 0.0250 | 0.0078 | 0.45 | 40.16 | <1.0 × 10−5 | B*38~C*12 | 0.0125 | 0.0056 | 1.00 | 51.20 | <1.0 × 10−5 |
| A*32~B*35 | 0.0225 | 0.0074 | 0.19 | 7.22 | 7.2 × 10−3 | B*42~C*17 | 0.0125 | 0.0056 | 1.00 | 179.06 | <1.0 × 10−5 |
| A*24~B*08 | 0.0200 | 0.0070 | 0.16 | 6.76 | 9.3 × 10−3 | B*15~C*02 | 0.0100 | 0.0050 | 0.31 | 39.12 | <1.0 × 10−5 |
| A*33~B*14 | 0.0200 | 0.0070 | 0.40 | 42.55 | <1.0 × 10−5 | B*45~C*16 | 0.0100 | 0.0050 | 1.00 | 55.82 | <1.0 × 10−5 |
| A*01~B*37 | 0.0150 | 0.0061 | 0.73 | 46.42 | <1.0 × 10−5 | B~DRB1 | |||||
| A*03~B*52 | 0.0150 | 0.0061 | 0.38 | 23.95 | <1.0 × 10−5 | B*08~DRB1*03 | 0.0975 | 0.0148 | 0.80 | 117.49 | <1.0 × 10−5 |
| A*03~B*18 | 0.0100 | 0.0050 | 0.25 | 9.46 | 2.1 × 10−3 | B*50~DRB1*07 | 0.0475 | 0.0106 | 0.51 | 64.97 | <1.0 × 10−5 |
| A*11~B*41 | 0.0100 | 0.0050 | 0.34 | 11.10 | 9.0 × 10−4 | B*52~DRB1*15 | 0.0300 | 0.0085 | 0.84 | 95.14 | <1.0 × 10−5 |
| B~C | B*40~DRB1*16 | 0.0275 | 0.0082 | 0.45 | 20.01 | 1.0 × 10−5 | |||||
| B*35~C*04 | 0.1175 | 0.0161 | 0.83 | 238.26 | <1.0 × 10−5 | B*58~DRB1*03 | 0.0250 | 0.0078 | 0.27 | 6.36 | 0.0117 |
| B*50~C*06 | 0.0800 | 0.0136 | 0.96 | 210.04 | <1.0 × 10−5 | B*14~DRB1*01 | 0.0150 | 0.0061 | 0.30 | 36.43 | <1.0 × 10−5 |
| B*51~C*15 | 0.0700 | 0.0128 | 0.43 | 44.92 | <1.0 × 10−5 | B*14~DRB1*07 | 0.0150 | 0.0061 | 0.24 | 7.18 | 7.4 × 10−3 |
| B*51~C*16 | 0.0525 | 0.0112 | 0.72 | 62.65 | <1.0 × 10−5 | B*18~DRB1*16 | 0.0150 | 0.0061 | 0.34 | 7.42 | 6.4 × 10−3 |
| B*14~C*08 | 0.0450 | 0.0104 | 1.00 | 400.00 | <1.0 × 10−5 | B*37~DRB1*10 | 0.0125 | 0.0056 | 0.61 | 78.06 | <1.0 × 10−5 |
| B*58~C*03 | 0.0450 | 0.0104 | 0.77 | 198.26 | <1.0 × 10−5 | B*41~DRB1*08 | 0.0100 | 0.0050 | 0.43 | 66.45 | <1.0 × 10−5 |
| B*40~C*15 | 0.0400 | 0.0098 | 0.77 | 83.56 | <1.0 × 10−5 | B*53~DRB1*04 | 0.0100 | 0.0050 | 0.38 | 11.70 | 6.0 × 10−4 |
| B*52~C*12 | 0.0325 | 0.0089 | 0.92 | 124.57 | <1.0 × 10−5 | B*49~DRB1*13 | 0.0100 | 0.0050 | 0.55 | 36.56 | <1.0 × 10−5 |
| B*51~C*14 | 0.0300 | 0.0085 | 1.00 | 51.07 | <1.0 × 10−5 | B*15~DRB1*11 | 0.0100 | 0.0050 | 0.28 | 13.18 | 3.0 × 10−4 |
| B*37~C*06 | 0.0200 | 0.0070 | 1.00 | 51.21 | <1.0 × 10−5 | B*42~DRB1*03 | 0.0100 | 0.0050 | 0.74 | 9.76 | 1.8 × 10−3 |
| B*73~C*15 | 0.0175 | 0.0066 | 1.00 | 47.68 | <1.0 × 10−5 | DRB1~DQB1 | |||||
| B*49~C*07 | 0.0175 | 0.0066 | 1.00 | 28.50 | <1.0 × 10−5 | DRB1*03~DQB1*02 | 0.2125 | 0.0205 | 0.93 | 204.69 | <1.0 × 10−5 |
| B*18~C*12 | 0.0175 | 0.0066 | 0.49 | 33.00 | <1.0 × 10−5 | DRB1*16~DQB1*05 | 0.1725 | 0.0189 | 0.96 | 187.82 | <1.0 × 10−5 |
| B*07~C*07 | 0.0150 | 0.0061 | 0.95 | 185.92 | <1.0 × 10−5 | DRB1*07~DQB1*02 | 0.1100 | 0.0157 | 0.79 | 76.03 | <1.0 × 10−5 |
| DRB1*04~DQB1*03 | 0.0875 | 0.0141 | 0.82 | 119.94 | <1.0 × 10−5 | DRB1*10~DQB1*05 | 0.0375 | 0.0095 | 1.00 | 36.80 | <1.0 × 10−5 |
| DRB1*15~DQB1*06 | 0.0850 | 0.0140 | 0.85 | 192.21 | <1.0 × 10−5 | DRB1*14~DQB1*05 | 0.0225 | 0.0074 | 1.00 | 21.74 | <1.0 × 10−5 |
| DRB1*11~DQB1*03 | 0.0650 | 0.0123 | 0.91 | 98.28 | <1.0 × 10−5 | DRB1*08~DQB1*04 | 0.0125 | 0.0056 | 0.54 | 80.11 | <1.0 × 10−5 |
| DRB1*13~DQB1*06 | 0.0475 | 0.0106 | 0.84 | 101.25 | <1.0 × 10−5 | DRB1*04~DQB1*04 | 0.0100 | 0.0050 | 0.22 | 6.15 | 0.0131 |
| DRB1*01~DQB1*05 | 0.0450 | 0.0104 | 1.00 | 44.51 | <1.0 × 10−5 | DRB1*12~DQB1*03 | 0.0100 | 0.0050 | 0.75 | 11.19 | 0.0008 |
| A~B~C~DRB1~DQB1 Haplotype | Frequency | Standard Deviation | A~B~C~DRB1~DQB1 Haplotype | Frequency | Standard Deviation |
|---|---|---|---|---|---|
| A*26~B*08~C*07~DRB1*03~DQB1*02 | 0.0425 | 0.0101 | A*11~B*51~C*07~DRB1*16~DQB1*05 | 0.0100 | 0.0050 |
| A*02~B*50~C*06~DRB1*07~DQB1*02 | 0.0150 | 0.0061 | A*32~B*08~C*07~DRB1*03~DQB1*02 | 0.0100 | 0.0050 |
| A*03~B*52~C*12~DRB1*15~DQB1*06 | 0.0150 | 0.0061 | A*01~B*37~C*06~DRB1*10~DQB1*05 | 0.0100 | 0.0050 |
| A*02~B*08~C*07~DRB1*03~DQB1*02 | 0.0150 | 0.0061 | A*11~B*41~C*07~DRB1*08~DQB1*03 | 0.0100 | 0.0050 |
| A*24~B*08~C*07~DRB1*03~DQB1*02 | 0.0150 | 0.0061 | A*03~B*50~C*06~DRB1*07~DQB1*02 | 0.0075 | 0.0043 |
| A*01~B*51~C*15~DRB1*03~DQB1*02 | 0.0125 | 0.0056 | A*68~B*35~C*04~DRB1*04~DQB1*03 | 0.0075 | 0.0043 |
| A*33~B*14~C*08~DRB1*01~DQB1*05 | 0.0125 | 0.0056 | A*03~B*50~C*06~DRB1*04~DQB1*04 | 0.0075 | 0.0043 |
| A*02~B*51~C*15~DRB1*04~DQB1*03 | 0.0125 | 0.0056 | A*01~B*58~C*03~DRB1*03~DQB1*02 | 0.0075 | 0.0043 |
| A*11~B*40~C*15~DRB1*16~DQB1*05 | 0.0125 | 0.0056 | A*26~B*51~C*14~DRB1*15~DQB1*06 | 0.0075 | 0.0043 |
| A*33~B*58~C*03~DRB1*03~DQB1*02 | 0.0100 | 0.0050 | A*11~B*40~C*15~DRB1*03~DQB1*02 | 0.0075 | 0.0043 |
| A*68 B*14 C*08 DRB1*07 DQB1*02 | 0.0100 | 0.0050 | A*32~B*18~C*12~DRB1*16~DQB1*05 | 0.0075 | 0.0043 |
| A*33 B*58 C*03 DRB1*16 DQB1*05 | 0.0100 | 0.0050 | A*02~B*51~C*16~DRB1*16~DQB1*05 | 0.0075 | 0.0043 |
| A*02 B*35 C*04 DRB1*16 DQB1*05 | 0.0100 | 0.0050 |
| HLA-A, -B, -DRB1, and -DQB1 (35 Populations) | HLA-DRB1 (85 Populations) | ||||
|---|---|---|---|---|---|
| Population | SGD * | Population | SGD | Population | SGD |
| Oman | 0.0276 | Emirates | 0.0012 | Italy | 0.2688 |
| Saudi Arabia-B | 0.0886 | Kuwait-A | 0.0456 | Spain-Basques-B | 0.2703 |
| Saudi Arabia-C | 0.0930 | Oman-A | 0.0589 | Greece-Cretans | 0.2738 |
| Saudi Arabia | 0.0982 | Bahrain | 0.0594 | Iran-A | 0.2799 |
| Tunisia-A | 0.1108 | Sardinia | 0.0622 | Iraq-kurdistan | 0.2899 |
| Algeria | 0.1127 | Khuzestani | 0.1068 | Jordan-A | 0.3086 |
| Spain | 0.1139 | Algeria-A | 0.1178 | Egypt-A | 0.3235 |
| Saudi Arabia-D | 0.1140 | Kuwait | 0.1186 | India-Northeast | 0.3269 |
| Tunisia-Gabes | 0.1246 | Tunisia-A | 0.1267 | Iran-Kurd | 0.3316 |
| Morocco | 0.1258 | Morocco-Agadir | 0.1310 | Palestine | 0.3322 |
| France | 0.1344 | Algeria | 0.1447 | Greece-D | 0.3470 |
| Pakistan | 0.1380 | Algeria | 0.1473 | Egyptians | 0.3498 |
| Macedonia | 0.1409 | Saudi Arabia-C | 0.1522 | Greece-C | 0.3568 |
| Greece-Cretans | 0.1436 | Saudi Arabia-B | 0.1530 | Ashkenazi-Jews | 0.3582 |
| Italy | 0.1442 | Gabesians | 0.1590 | Syria-A | 0.3704 |
| Tunisia | 0.1530 | Tunisia | 0.1643 | Syria | 0.3799 |
| South Tunisia | 0.1569 | Saudi Arabia-D | 0.1716 | Albania | 0.3813 |
| Iraq-Kurdis | 0.1598 | Morocco-A | 0.1747 | Sudan | 0.3844 |
| Saudi Arabia-A | 0.1732 | Spain-Spaniards | 0.1770 | Greece-A | 0.3944 |
| Greece-D | 0.1786 | Saudi Aarbia | 0.1780 | Iran-Azeri | 0.4072 |
| Spain-Basques | 0.1883 | Southern Tunisia | 0.1817 | Moroccans-Jews | 0.4130 |
| Tunisia-Ghannouch | 0.1917 | Tunisia- Berbers-Met | 0.1914 | Lebanon-Yohmor | 0.4397 |
| Palestine | 0.2118 | Portugal | 0.1995 | Sri lanka | 0.4477 |
| Greece-A | 0.2213 | Morocco | 0.2001 | Libya-Jews | 0.4537 |
| Spain-Basques | 0.2226 | Libya | 0.2022 | Lebanon-B | 0.4603 |
| Ashkenazi-j | 0.2264 | France-Rennes | 0.2024 | Thailland | 0.4689 |
| Morocco-Jews | 0.2450 | Basques-A | 0.2053 | Lebanon-Armen | 0.4896 |
| Turkey-A | 0.2564 | France | 0.2147 | Bangladesh | 0.4928 |
| Spain-Berbers | 0.2775 | Tunisia-Berbers | 0.2169 | Turkey-A | 0.5012 |
| Lebanon-A | 0.2817 | Iran-Famoori | 0.2170 | Lebanon-A | 0.5068 |
| Guinea-Bubi | 0.4143 | Morocco-Chaouya | 0.2170 | Lebanon-KZ | 0.5378 |
| Senegal-Mandenka | 0.4218 | Yemen-sana’a | 0.2278 | Lebanon | 0.5956 |
| Mali-Fulani | 0.4509 | Pakistan-Sindh | 0.2284 | Senegal | 0.6885 |
| Mali- Mossi | 0.5496 | Tunisia-Ghannouchians | 0.2292 | Senegal-Mandenka | 0.7174 |
| Mali-Rimaibe | 0.6432 | Macedonia | 0.2314 | Iran-Jews | 0.7476 |
| Basques-Arratia | 0.2390 | Lebanon-NS | 0.7612 | ||
| Turkey | 0.2394 | Guinia-Bubi | 0.7675 | ||
| Pakistan-Pathan | 0.2414 | Indonesia | 0.9049 | ||
| India-Delhi | 0.2465 | Mali-Fulani | 1.0674 | ||
| Yemen-Jews | 0.2533 | Mali-Mossi | 1.3297 | ||
| Murcians | 0.2616 | Rimaibe | 1.4181 | ||
| Greeks-B | 0.2642 | ||||
| Iranians | 0.2688 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Yafei, Z.; Hajjej, A.; Alvares, M.; Al Mahri, A.; Nasr, A.; Mirghani, R.; Al Obaidli, A.; Al Seiari, M.; Mack, S.J.; Askar, M.; et al. Analysis of the Origin of Emiratis as Inferred from a Family Study Based on HLA-A, -C, -B, -DRB1, and -DQB1 Genes. Genes 2023, 14, 1159. https://doi.org/10.3390/genes14061159
Al Yafei Z, Hajjej A, Alvares M, Al Mahri A, Nasr A, Mirghani R, Al Obaidli A, Al Seiari M, Mack SJ, Askar M, et al. Analysis of the Origin of Emiratis as Inferred from a Family Study Based on HLA-A, -C, -B, -DRB1, and -DQB1 Genes. Genes. 2023; 14(6):1159. https://doi.org/10.3390/genes14061159
Chicago/Turabian StyleAl Yafei, Zain, Abdelhafidh Hajjej, Marion Alvares, Ayeda Al Mahri, Amre Nasr, Rajaa Mirghani, Ali Al Obaidli, Mohamed Al Seiari, Steven J. Mack, Medhat Askar, and et al. 2023. "Analysis of the Origin of Emiratis as Inferred from a Family Study Based on HLA-A, -C, -B, -DRB1, and -DQB1 Genes" Genes 14, no. 6: 1159. https://doi.org/10.3390/genes14061159
APA StyleAl Yafei, Z., Hajjej, A., Alvares, M., Al Mahri, A., Nasr, A., Mirghani, R., Al Obaidli, A., Al Seiari, M., Mack, S. J., Askar, M., Edinur, H. A., Almawi, W. Y., & ElGhazali, G. (2023). Analysis of the Origin of Emiratis as Inferred from a Family Study Based on HLA-A, -C, -B, -DRB1, and -DQB1 Genes. Genes, 14(6), 1159. https://doi.org/10.3390/genes14061159






