Integration of Phosphoproteomics and Transcriptome Studies Reveals ABA Signaling Pathways Regulate UV-B Tolerance in Rhododendron chrysanthum Leaves
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Determination of Stomatal Characteristics
2.3. RNA-Seq Library Construction and Sequencing
2.4. De Novo Assembly and Sequence Annotation
2.5. Differential Expression Analysis
2.6. Protein Extraction and Digestion
2.7. Phosphopeptide Enrichment
2.8. LC-MS/MS Analysis and Database Search
2.9. Bioinformatics
3. Results
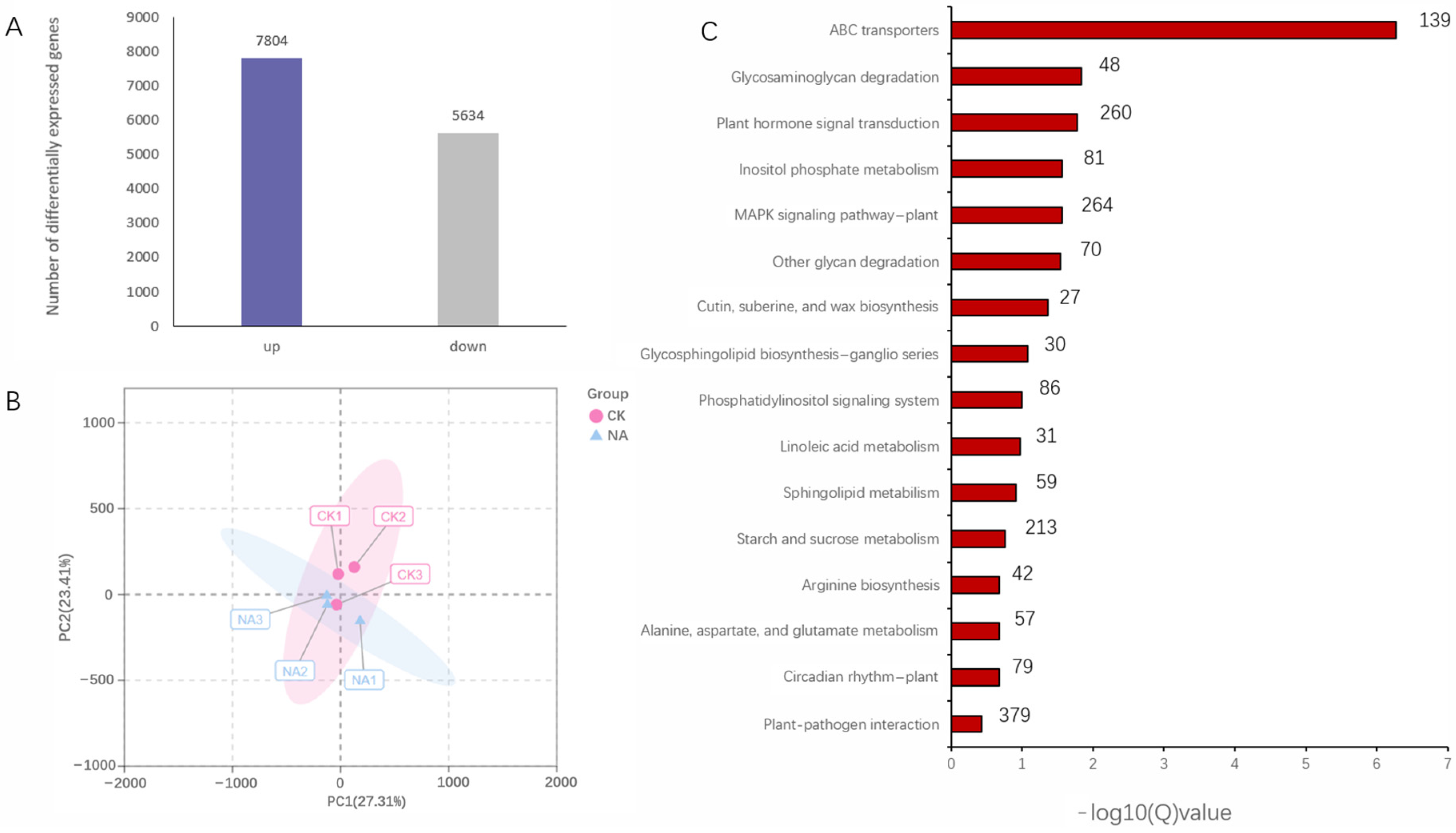
3.1. Transcriptomic Analysis of R. chrysanthum under UV-B Stress
3.2. Phosphorylated Proteomics Analysis of R. chrysanthum under UV-B Stress
3.3. Three-Dimensional Structure Modeling of UV-B Stress-Responsive Phosphoproteins
3.4. Transcriptomic and Phosphorylated Proteomic Interaction Analysis to Explore the Response of R. chrysanthum to UV-B Stress
3.5. Effect of Exogenous ABA on Stomata of R. chrysanthum after UV-B Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Rodrigues, A.; Pizzio, G.A.; Rodriguez, P.L. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 2012, 158, 970–980. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xu, Z.; Fan, X.; Zhou, Q.; Cao, J.; Ji, G.; Jing, S.; Feng, B.; Wang, T. Transcriptome Analysis Reveals Complex Molecular Mechanisms Underlying UV Tolerance of Wheat (Triticum aestivum, L.). J. Agric. Food Chem. 2019, 67, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, Q.; Cao, K.; Xu, H.; Zhou, X. Acetylated Proteomics of UV-B Stress-Responsive in Photosystem II of Rhododendron chrysanthum. Cells 2023, 12, 478. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Liu, M.; Cao, K.; Xu, H.; Zhou, X. UV-B Irradiation to Amino Acids and Carbohydrate Metabolism in Rhododendron chrysanthum Leaves by Coupling Deep Transcriptome and Metabolome Analysis. Plants 2022, 11, 2730. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Sun, L.; Chu, S.; Xu, H.; Zhou, X. Integration of transcriptomic and proteomic analyses of Rhododendron chrysanthum Pall. in response to cold stress in the Changbai Mountains. Mol. Biol. Rep. 2023, 50, 3607–3616. [Google Scholar] [CrossRef]
- Bharath, P.; Gahir, S.; Raghavendra, A.S. Abscisic Acid-Induced Stomatal Closure: An Important Component of Plant Defense Against Abiotic and Biotic Stress. Front. Plant Sci. 2021, 12, 615114. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in Stomatal Defense against Biotic and Drought Stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Niu, M.; Xie, J.; Chen, C.; Cao, H.; Sun, J.; Kong, Q.; Shabala, S.; Shabala, L.; Huang, Y.; Bie, Z. An early ABA-induced stomatal closure, Na+ sequestration in leaf vein and K+ retention in mesophyll confer salt tissue tolerance in Cucurbita species. J. Exp. Bot. 2018, 69, 4945–4960. [Google Scholar] [CrossRef]
- Li, Z.; Takahashi, Y.; Scavo, A.; Brandt, B.; Nguyen, D.; Rieu, P.; Schroeder, J.I. Abscisic acid-induced degradation of Arabidopsis guanine nucleotide exchange factor requires calcium-dependent protein kinases. Proc. Natl. Acad. Sci. USA 2018, 115, E4522–E4531. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- García-León, M.; Cuyas, L.; El-Moneim, D.A.; Rodriguez, L.; Belda-Palazón, B.; Sanchez-Quant, E.; Fernández, Y.; Roux, B.; Zamarreño, Á.M.; García-Mina, J.M.; et al. Arabidopsis ALIX Regulates Stomatal Aperture and Turnover of Abscisic Acid Receptors. Plant Cell 2019, 31, 2411–2429. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate—Stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef] [PubMed]
- Hewage, K.A.H.; Yang, J.F.; Wang, D.; Hao, G.F.; Yang, G.F.; Zhu, J.K. Chemical Manipulation of Abscisic Acid Signaling: A New Approach to Abiotic and Biotic Stress Management in Agriculture. Adv. Sci. 2020, 7, 2001265. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, C.; Kong, D.; Guo, F.; Wei, H. Light-Mediated Signaling and Metabolic Changes Coordinate Stomatal Opening and Closure. Front. Plant Sci. 2020, 11, 601478. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.M.; Ishikawa, S.; Takahashi, F.; Suzuki, K.; Kamo, M.; Umezawa, T.; Shinozaki, K.; Kawamura, Y.; Uemura, M. Large-Scale Phosphoproteomic Study of Arabidopsis Membrane Proteins Reveals Early Signaling Events in Response to Cold. Int. J. Mol. Sci. 2020, 21, 8631. [Google Scholar] [CrossRef] [PubMed]
- Rampitsch, C.; Bykova, N.V. The beginnings of crop phosphoproteomics: Exploring early warning systems of stress. Front. Plant Sci. 2012, 3, 144. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, H.; Dong, J.; Chen, J.; Xu, H.; Zhou, X. Phosphoproteomics of cold stress-responsive mechanisms in Rhododendron chrysanthum. Mol. Biol. Rep. 2022, 49, 303–312. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Cao, K.; Xu, H.; Zhou, X. Transcriptome and proteome depth analysis indicate ABA, MAPK cascade and Ca(2+) signaling co-regulate cold tolerance in Rhododendron chrysanthum Pall. Front. Plant Sci. 2023, 14, 1146663. [Google Scholar] [CrossRef]
- Lyu, J.; Wang, C.; Liang, D.Y.; Liu, L.; Zhou, X.F. Sensitivity of wild and domesticated Rhododendron chrysanthum to different light regime (UVA, UVB, and PAR). Photosynthetica 2020, 57, 841–849. [Google Scholar] [CrossRef]
- Zhou, X.; Lyu, J.; Sun, L.; Dong, J.; Xu, H. Metabolic programming of Rhododendron chrysanthum leaves following exposure to UVB irradiation. Funct. Plant Biol. 2021, 48, 1175–1185. [Google Scholar] [CrossRef]
- Czégény, G.; Mátai, A.; Hideg, É. UV-B effects on leaves-Oxidative stress and acclimation in controlled environments. Plant Sci. Int. J. Exp. Plant Biol. 2016, 248, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Millstead, L.; Jayakody, H.; Patel, H.; Kaura, V.; Petrie, P.R.; Tomasetig, F.; Whitty, M. Accelerating Automated Stomata Analysis Through Simplified Sample Collection and Imaging Techniques. Front. Plant Sci. 2020, 11, 580389. [Google Scholar] [CrossRef] [PubMed]
- Rha, E.Y.; Kim, J.M.; Yoo, G. Volume Measurement of Various Tissues Using the Image J Software. J. Craniofacial Surg. 2015, 26, e505–e506. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Zhang, Z.; Fan, H.; Tan, Y.; Xu, H.; Zhou, X. Comparative transcriptomic analysis reveals gene expression in response to cold stress in Rhododendron aureum Georgi. Theor. Exp. Plant Physiol. 2022, 34, 347–366. [Google Scholar] [CrossRef]
- Beltran, L.; Cutillas, P.R. Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids 2012, 43, 1009–1024. [Google Scholar] [CrossRef]
- Paulo, J.A.; Schweppe, D.K. Advances in quantitative high-throughput phosphoproteomics with sample multiplexing. Proteomics 2021, 21, e2000140. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Li, S.; Deng, B.; Tian, S.; Guo, M.; Liu, H.; Zhao, X. Metabolic and transcriptomic analyses reveal different metabolite biosynthesis profiles between leaf buds and mature leaves in Ziziphus jujuba mill. Food Chem. 2021, 347, 129005. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Thakur, P.; Kumar, S.; Malik, J.A.; Berger, J.D.; Nayyar, H. Cold stress effects on reproductive development in grain crops: An overview. Environ. Exp. Bot. 2010, 67, 429–443. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Y.; Zhang, Z.; Liu, X.; Hsu, C.C.; Du, Y.; Sang, T.; Zhu, C.; Wang, Y.; Satheesh, V.; et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 2020, 11, 613. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e5. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Grennan, A.K. Protein S-nitrosylation: Potential targets and roles in signal transduction. Plant Physiol. 2007, 144, 1237–1239. [Google Scholar] [CrossRef]
- Pi, Z.; Zhao, M.L.; Peng, X.J.; Shen, S.H. Phosphoproteomic Analysis of Paper Mulberry Reveals Phosphorylation Functions in Chilling Tolerance. J. Proteome Res. 2017, 16, 1944–1961. [Google Scholar] [CrossRef]
- Lv, D.W.; Subburaj, S.; Cao, M.; Yan, X.; Li, X.; Appels, R.; Sun, D.F.; Ma, W.; Yan, Y.M. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell. Proteom. MCP 2014, 13, 632–652. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, C.Y.; Lv, D.W.; Zhen, S.M.; Li, X.H.; Yan, Y.M. Comparative phosphoproteome analysis of the developing grains in bread wheat (Triticum aestivum L.) under well-watered and water-deficit conditions. J. Proteome Res. 2014, 13, 4281–4297. [Google Scholar] [CrossRef]
- Bögre, L.; Ligterink, W.; Heberle-Bors, E.; Hirt, H. Mechanosensors in plants. Nature 1996, 383, 489–490. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Zhang, Y.; Ren, H. Controlling the Gate: The Functions of the Cytoskeleton in Stomatal Movement. Front. Plant Sci. 2022, 13, 849729. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Q.; Zhou, X.; Yang, L.; Xu, H.; Zhou, X. Integration of Phosphoproteomics and Transcriptome Studies Reveals ABA Signaling Pathways Regulate UV-B Tolerance in Rhododendron chrysanthum Leaves. Genes 2023, 14, 1153. https://doi.org/10.3390/genes14061153
Sun Q, Zhou X, Yang L, Xu H, Zhou X. Integration of Phosphoproteomics and Transcriptome Studies Reveals ABA Signaling Pathways Regulate UV-B Tolerance in Rhododendron chrysanthum Leaves. Genes. 2023; 14(6):1153. https://doi.org/10.3390/genes14061153
Chicago/Turabian StyleSun, Qi, Xiangru Zhou, Liping Yang, Hongwei Xu, and Xiaofu Zhou. 2023. "Integration of Phosphoproteomics and Transcriptome Studies Reveals ABA Signaling Pathways Regulate UV-B Tolerance in Rhododendron chrysanthum Leaves" Genes 14, no. 6: 1153. https://doi.org/10.3390/genes14061153
APA StyleSun, Q., Zhou, X., Yang, L., Xu, H., & Zhou, X. (2023). Integration of Phosphoproteomics and Transcriptome Studies Reveals ABA Signaling Pathways Regulate UV-B Tolerance in Rhododendron chrysanthum Leaves. Genes, 14(6), 1153. https://doi.org/10.3390/genes14061153






