Evaluating the Differential Response of Transcription Factors in Diploid versus Autotetraploid Rice Leaves Subjected to Diverse Saline–Alkali Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatments
2.2. RNA Isolation and RNA-seq
2.3. Data Analysis
2.4. qRT-PCR Validation
3. Results
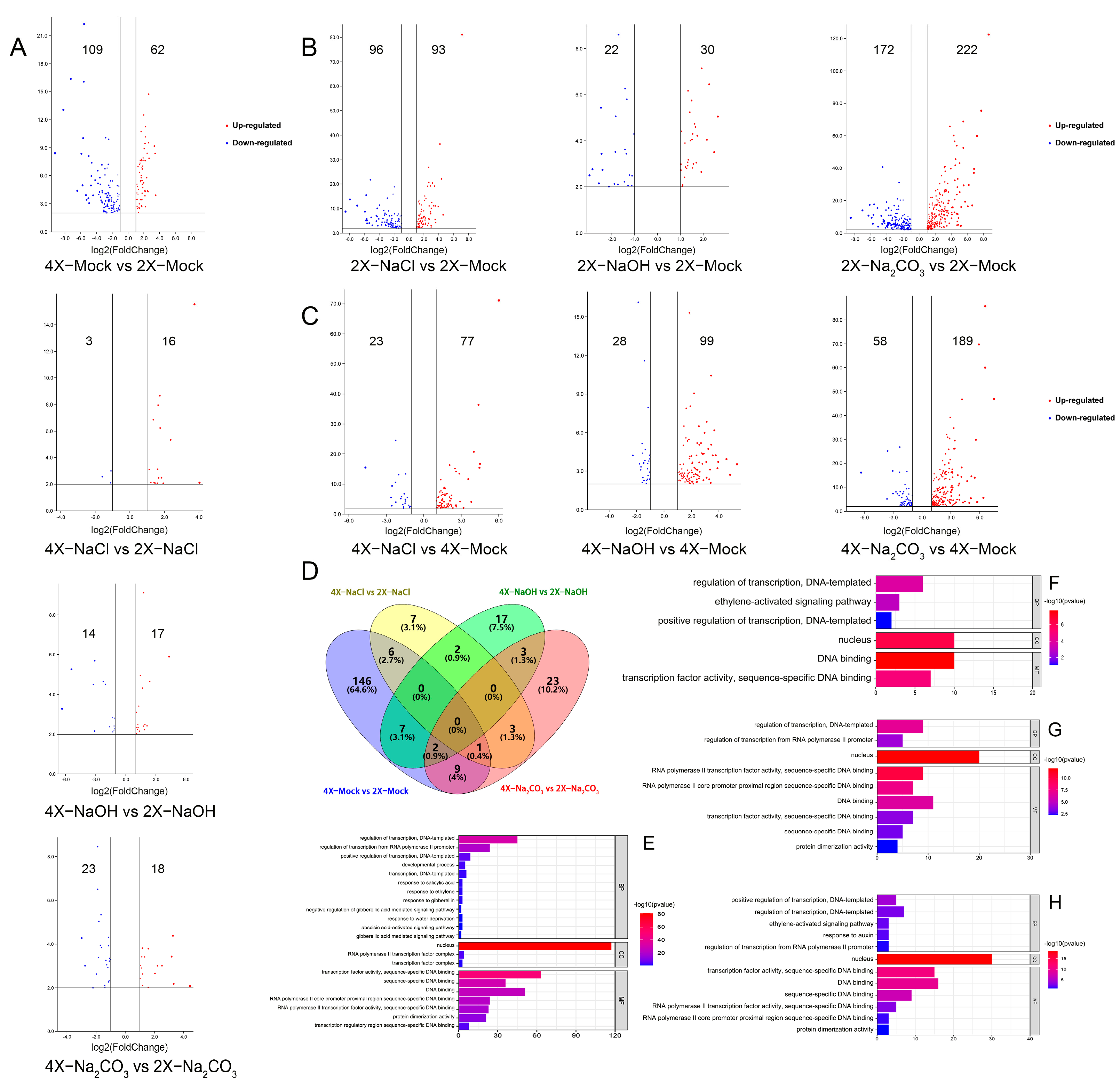
3.1. Genome-Wide Expression of TFs-Related Genes Varied between 9311-2x and 9311-4x Rice under Saline–Alkali Stress
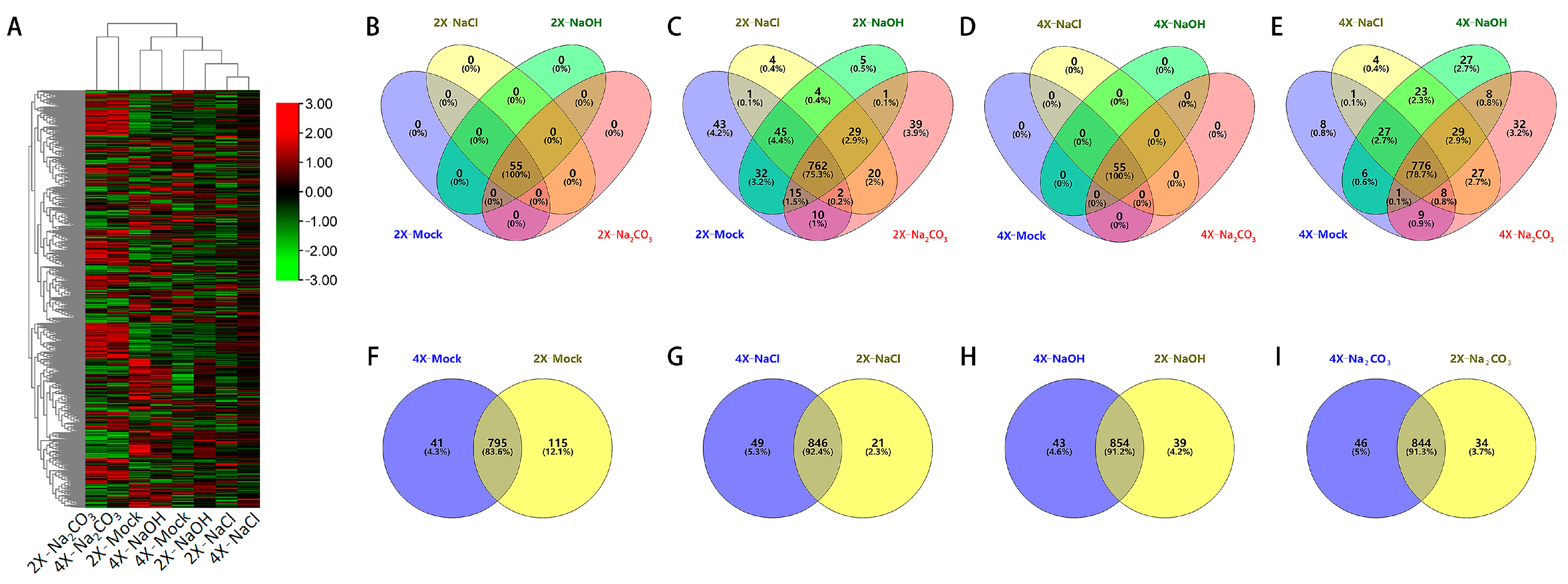
3.2. Analysis of Differentially Expressed 56 Transcription Factor Families’ Genes in Diploid and Autotetraploid Rice Leaves
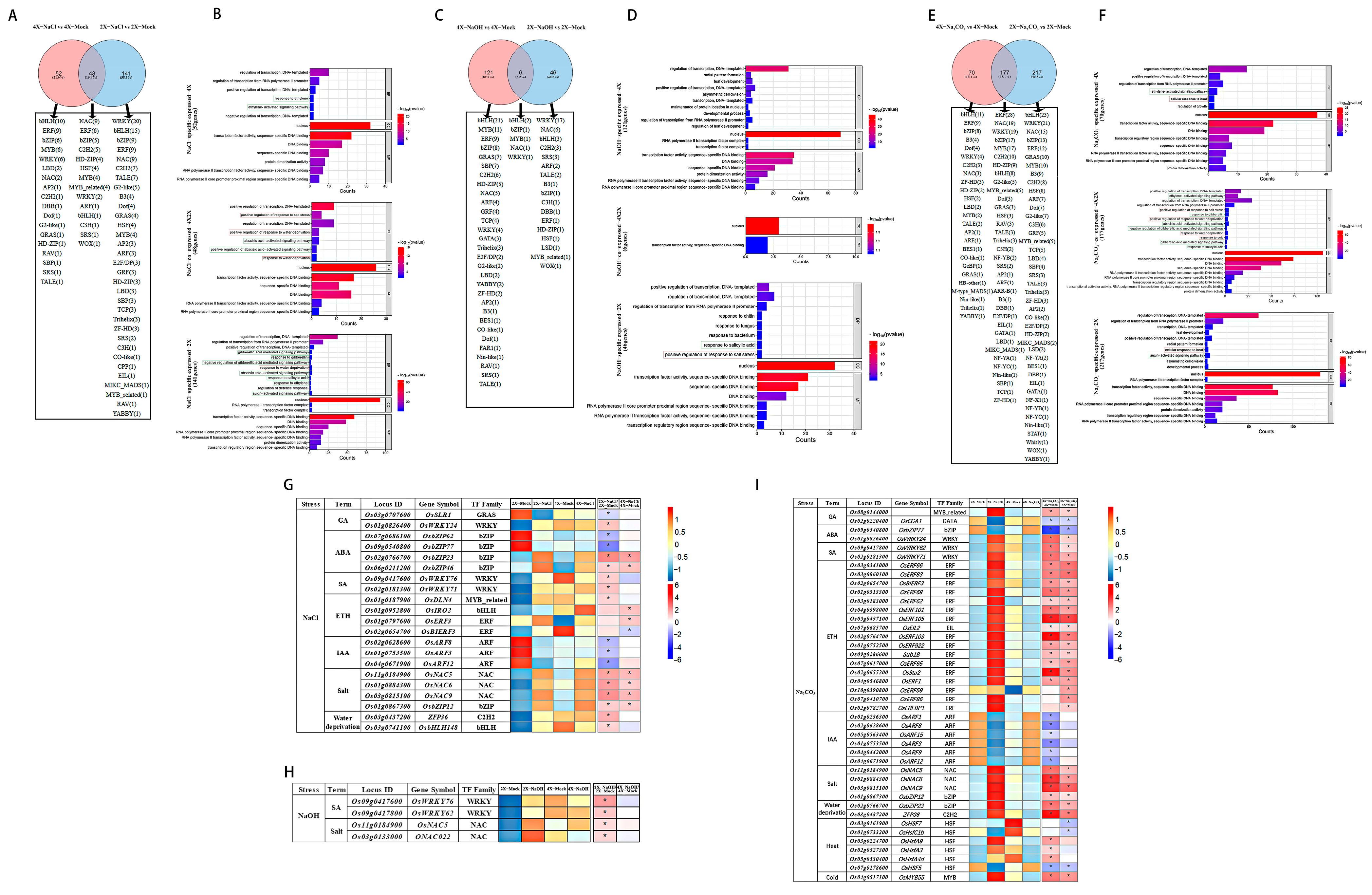
3.3. Comparative Analyses of the DETF Genes in Rice Induced by Salt, Alkali, and Saline–Alkali Stress Treatments
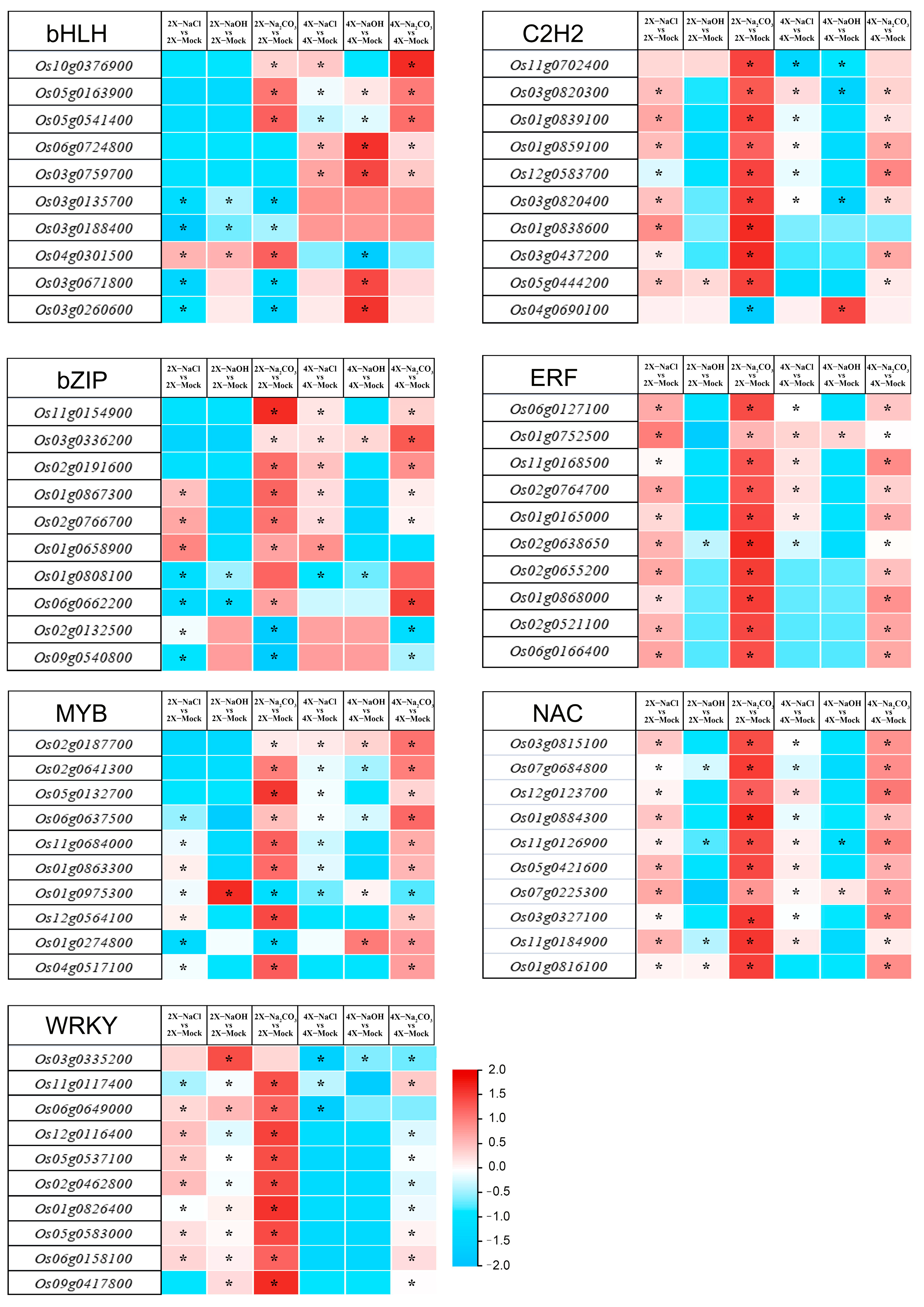
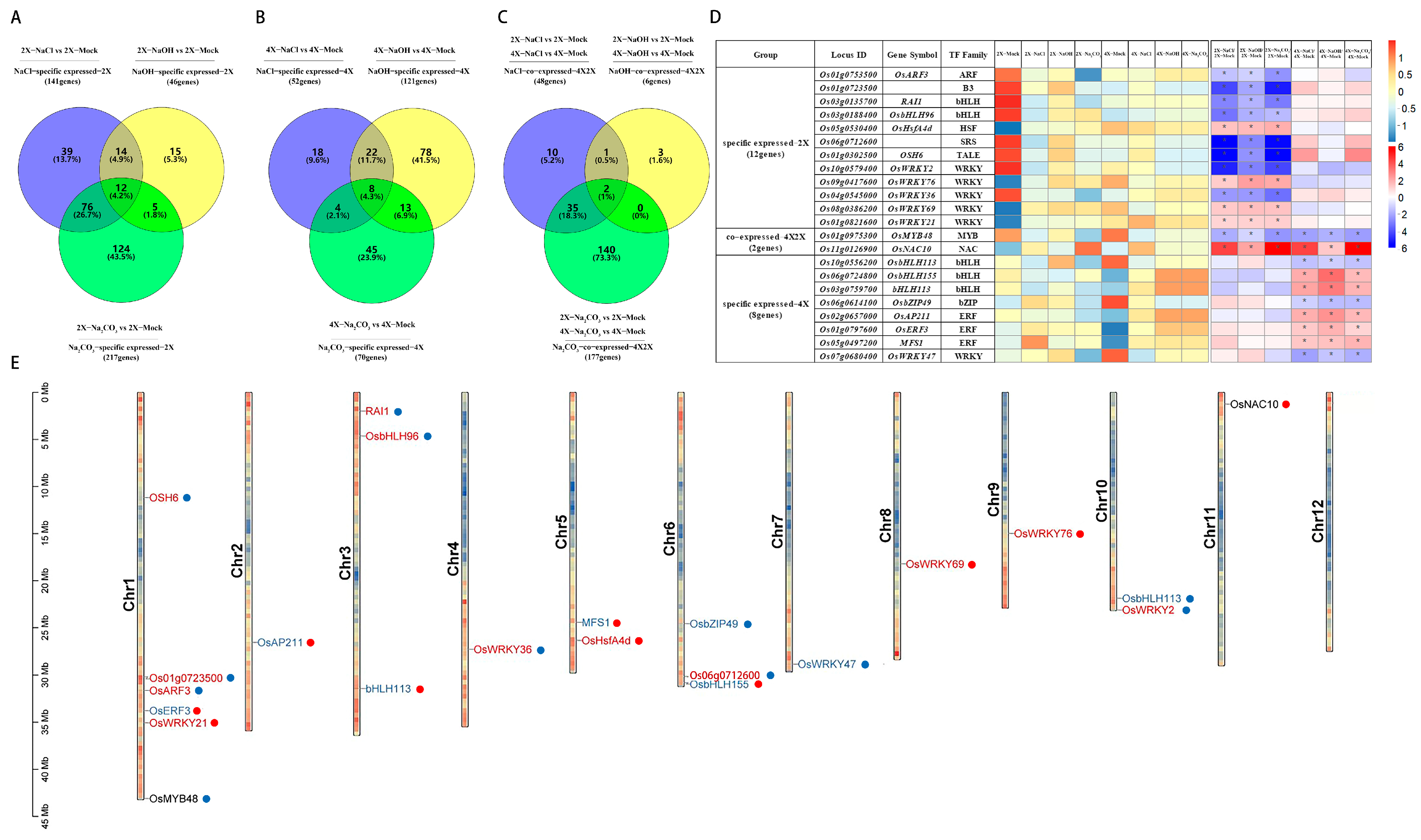
3.4. Identification of the Common Expressed DETF Genes in the 9311-2x and 9311-4x Leaves in Response to Salt, Alkali, and Saline–Alkali Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Li, S.; Tian, S.; Wang, B.; Zhao, X. Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube (Ziziphus acidojujuba). PLoS ONE 2017, 12, e0185732. [Google Scholar] [CrossRef]
- Cao, Y.; Song, H.; Zhang, L. New Insight into Plant Saline-Alkali Tolerance Mechanisms and Application to Breeding. Int. J. Mol. Sci. 2022, 23, 16048. [Google Scholar] [CrossRef]
- Oster, J.; Shainberg, I.; Abrol, I. Reclamation of salt-affected soils. Agric. Drain 1999, 38, 659–691. [Google Scholar]
- Tossi, V.E.; Martinez Tosar, L.J.; Laino, L.E.; Iannicelli, J.; Regalado, J.J.; Escandon, A.S.; Baroli, I.; Causin, H.F.; Pitta-Alvarez, S.I. Impact of polyploidy on plant tolerance to abiotic and biotic stresses. Front. Plant Sci. 2022, 13, 869423. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Ashman, T.L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef]
- Xiao, L.; Lu, L.; Zeng, W.; Shang, X.; Cao, S.; Yan, H. DNA Methylome and LncRNAome Analysis Provide Insights into Mechanisms of Genome-Dosage Effects in Autotetraploid Cassava. Front. Plant Sci. 2022, 13, 915056. [Google Scholar] [CrossRef]
- Liu, D.; Cheng, Y.; Gong, M.; Zhao, Q.; Jiang, C.; Cheng, L.; Ren, M.; Wang, Y.; Yang, A. Comparative transcriptome analysis reveals differential gene expression in resistant and susceptible tobacco cultivars in response to infection by cucumber mosaic virus. Crop J. 2019, 7, 307–321. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J.K. Understanding and improving salt tolerance in plants. Crop Sci. 2005, 45, 437–448. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, W.; Wang, Y.; Zhou, Y.; Wang, S.; Qi, F.; Wang, N.; Ma, J. Comparative Analysis of Physiological, Hormonal and Transcriptomic Responses Reveal Mechanisms of Saline-Alkali Tolerance in Autotetraploid Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 16146. [Google Scholar] [CrossRef]
- Wang, N.; Wang, S.; Qi, F.; Wang, Y.; Lin, Y.; Zhou, Y.; Meng, W.; Zhang, C.; Wang, Y.; Ma, J. Autotetraploidization Gives Rise to Differential Gene Expression in Response to Saline Stress in Rice. Plants 2022, 11, 3114. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Fan, X.; Lin, Y.; Li, Z.; Wang, Y.; Zhou, Y.; Meng, W.; Peng, Z.; Zhang, C.; Ma, J. Alkaline Stress Induces Different Physiological, Hormonal and Gene Expression Responses in Diploid and Autotetraploid Rice. Int. J. Mol. Sci. 2022, 23, 5561. [Google Scholar] [CrossRef]
- Koide, Y.; Kuniyoshi, D.; Kishima, Y. Fertile Tetraploids: New Resources for Future Rice Breeding? Front. Plant Sci. 2020, 11, 1231. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, H.; Jin, J.; Ma, X.; Li, K. Physiological and Transcriptome Analysis on Diploid and Polyploid Populus ussuriensis Kom. under Salt Stress. Int. J. Mol. Sci. 2022, 23, 7529. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Zou, L.; Li, T.; Li, B.; He, J.; Liao, C.; Wang, L.; Xue, S.; Sun, T.; Ma, X.; Wu, Q. De novo transcriptome analysis provides insights into the salt tolerance of Podocarpus macrophyllus under salinity stress. BMC Plant Biol. 2021, 21, 489. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, H.; Ren, J.; Dong, J.; Zhao, X.; Wang, X.; Wang, J.; Zhong, C.; Zhao, S.; Liu, X.; et al. Comparative Transcriptome-Based Mining and Expression Profiling of Transcription Factors Related to Cold Tolerance in Peanut. Int. J. Mol. Sci. 2020, 21, 1921. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, X.; Xu, C.; Guo, D.; Niu, L.; Wang, Y.; Li, J.; Yan, F.; Wang, Q. Overexpression of a novel transcriptional repressor GmMYB3a negatively regulates salt-alkali tolerance and stress-related genes in soybean. Biochem. Biophys. Res. Commun. 2018, 498, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Show, P.L.; Miao, X. Transcription Factor ChbZIP1 from Alkaliphilic Microalgae Chlorella sp. BLD Enhancing Alkaline Tolerance in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 2387. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Baoxiang, W.; Jingfang, L.; Zhiguang, S.; Ming, C.; Yungao, X.; Bo, X.; Bo, Y.; Jian, L.; Jinbo, L.; et al. A novel SAPK10-WRKY87-ABF1 biological pathway synergistically enhance abiotic stress tolerance in transgenic rice (Oryza sativa). Plant Physiol. Biochem. 2021, 168, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, H.; Sun, C.; Ma, Q.; Bu, H.; Chong, K.; Xu, Y. A C(2)H(2) zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 2018, 229, 100–110. [Google Scholar] [CrossRef]
- Xu, G.; Cui, Y.; Wang, M.; Li, M.; Yin, X.; Xia, X. OsMsr9, a novel putative rice F-box containing protein, confers enhanced salt tolerance in transgenic rice and Arabidopsis. Molecular Breeding 2014, 34, 1055–1064. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Droge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Sun, B.; He, H.; Zhang, Y.; Tian, H.; Wang, B. Current Understanding of bHLH Transcription Factors in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 2015, 16, 227. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Y.; Liu, K.; Wang, N.; Wu, Y.; Zhang, C.; Ma, J. Transcriptome-Based Comparative Analysis of Transcription Factors in Response to NaCl, NaOH, and Na2CO3 Stresses in Roots of Autotetraploid Rice (Oryza sativa L.). Agronomy 2023, 13, 959. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Wang, Y.; Wang, C.; Leng, Z.; Qi, F.; Wang, S.; Zhou, Y.; Meng, W.; Liu, K.; Zhang, C.; et al. Evaluating the Differential Response of Transcription Factors in Diploid versus Autotetraploid Rice Leaves Subjected to Diverse Saline–Alkali Stresses. Genes 2023, 14, 1151. https://doi.org/10.3390/genes14061151
Wang N, Wang Y, Wang C, Leng Z, Qi F, Wang S, Zhou Y, Meng W, Liu K, Zhang C, et al. Evaluating the Differential Response of Transcription Factors in Diploid versus Autotetraploid Rice Leaves Subjected to Diverse Saline–Alkali Stresses. Genes. 2023; 14(6):1151. https://doi.org/10.3390/genes14061151
Chicago/Turabian StyleWang, Ningning, Yingkai Wang, Chenxi Wang, Zitian Leng, Fan Qi, Shiyan Wang, Yiming Zhou, Weilong Meng, Keyan Liu, Chunying Zhang, and et al. 2023. "Evaluating the Differential Response of Transcription Factors in Diploid versus Autotetraploid Rice Leaves Subjected to Diverse Saline–Alkali Stresses" Genes 14, no. 6: 1151. https://doi.org/10.3390/genes14061151
APA StyleWang, N., Wang, Y., Wang, C., Leng, Z., Qi, F., Wang, S., Zhou, Y., Meng, W., Liu, K., Zhang, C., & Ma, J. (2023). Evaluating the Differential Response of Transcription Factors in Diploid versus Autotetraploid Rice Leaves Subjected to Diverse Saline–Alkali Stresses. Genes, 14(6), 1151. https://doi.org/10.3390/genes14061151





