Abstract
The tonoplast monosaccharide transporter (TMT) family plays essential roles in sugar transport and plant growth. However, there is limited knowledge about the evolutionary dynamics of this important gene family in important Gramineae crops and putative function of rice TMT genes under external stresses. Here, the gene structural characteristics, chromosomal location, evolutionary relationship, and expression patterns of TMT genes were analyzed at a genome-wide scale. We identified six, three, six, six, four, six, and four TMT genes, respectively, in Brachypodium distachyon (Bd), Hordeum vulgare (Hv), Oryza rufipogon (Or), Oryza sativa ssp. japonica (Os), Sorghum bicolor (Sb), Setaria italica (Si), and Zea mays (Zm). All TMT proteins were divided into three clades based on the phylogenetic tree, gene structures, and protein motifs. The transcriptome data and qRT-PCR experiments suggested that each clade members had different expression patterns in various tissues and multiple reproductive tissues. In addition, the microarray datasets of rice indicated that different rice subspecies responded differently to the same intensity of salt or heat stress. The Fst value results indicated that the TMT gene family in rice was under different selection pressures in the process of rice subspecies differentiation and later selection breeding. Our findings pave the way for further insights into the evolutionary patterns of the TMT gene family in the important Gramineae crops and provide important references for characterizing the functions of rice TMT genes.
1. Introduction
In plants, sugars (including sucrose, monosaccharide, and polyols) participate in plant growth, development, and fruit flavor [1,2]. To date, various types of sugar transporters that transport polyols [3,4], monosaccharides [5], or sucrose [6,7] have been reported, such as the sugars will eventually be exported transporters (SWEET), sucrose transporters (SUT), and monosaccharide transporters (MST) gene families. Among them, the TMT proteins localizing on the tonoplast belong to the MST gene family. So far, genome-wide analyses have identified three, three, and six TMT genes in Arabidopsis, rice, and Pyrus bretschneideri, respectively [8,9,10].
The protein structures of the TMT gene family members are different from other monosaccharide transporter subfamilies (e.g., STP, PLT, ERD, pGlcT, INT, and XTPH). TMT proteins have a large cytoplasmic ring consisting of about 170 amino terminals between the 6th and 7th transmembrane domains. A large number of phosphorylation sites exist in this large cytoplasmic ring, which are similar to yeast glucose sensors, SNF, and RGT2. Therefore, it has been speculated that TMT protein has a sugar sensing function [11]. Vacuoles from Arabidopsis attmt1-2-3 (AtTMT1, -2, and -3 knockout lines) have shown an evident decrease in the absorption capacity of glucose [8]. Expressing the OsTMT1 gene in vacuoles of the mutant Arabidopsis (attmt1-2) has demonstrated that OsTMT proteins were capable of transporting glucose into vacuoles [9]. In addition, Arabidopsis mutant (attmt1/attmt2) has shown significant decreases in sucrose absorption capacity in vacuoles, which evidenced that AtTMT1 and AtTMT2 may also be involved in sucrose transportation across vacuole membranes [12]. The expression patterns of TMT genes in Arabidopsis show tissue-specific expression. Among them, AtTMT1 is mainly expressed in leaves and flowers. AtTMT2 has been shown to be more dominant in stems and roots, while AtTMT3 was weakly expressed in all tested tissues [12]. In rice, the expression patterns of OsTMT1 and OsTMT2 were very similar and showed an overlapping expression pattern. These two TMT genes have high expression levels in rice microtubule sheath cells, parenchyma cells of leaves, and companion cells [8,9]. Most PbTMTs are expressed to varying degrees in all tissues; however, PbTMT5 is highly expressed in stems and flowers [10]. In addition, PbTMT1 shows relatively uniform expression in various tissues, while PbTMT2, PbTMT3, and PbTMT6 are highly expressed in mature leaves. PbTMT4 is highly expressed in ripe fruit, possibly suggesting a key role in fruit sugar accumulation [10]. The over-expressing AtTMT1 in Arabidopsis has exhibited high growth rate at the seedling stage and increased seed biomass [13]. Wingenter et al. (2010) explained that these physiological changes were due to overexpression of the TMT gene, resulting in more efficient sugar sensing amplification and assimilate distribution in plants [13]. In addition, the expression of AtTMT1 and AtTMT2 have also been induced by drought, salinity, and chilling stresses [8,11].
The Gramineae class contains a variety of important food crops, with wide distribution areas, which play crucial roles in the global food supply [1,14]. With the development of sequencing technology, whole genome sequencing of rice, wild rice, barley, sorghum, millet, and maize in the Gramineae class have been completed, which provides basic genome data for studying important gene families at the genome-wide level. Based on the public genome sequences of seven Gramineae crops, in this study, we identified the TMT genes in B. distachyon (Bd) [15], H. vulgare (Hv) [16], S. italica (Si) [17], S. bicolor (Sb) [18], Z. mays (Zm) [19], O. rufipogon (Or) [20], and O. sativa ssp. japonica (Os) genomes [21,22]. Subsequently, we analyzed the gene structural features, chromosomal locations, and evolutionary relationships of TMT genes at a genome-wide scale, and analyzed the expression profiles of rice TMT genes in different tissues at different developmental stages or under different abiotic stresses. The current study provides a theoretical basis for exploring structure characteristics, putative functions, and evolutionary relationships of the TMT genes in these important Gramineae crops.
2. Materials and Methods
2.1. Plant Materials and Treatments
Fifteen rice tissues were collected from “9311” plants grown in a natural environment, during the summer, in Wuhan city (29°58′20″ N, 113°53′29″ E), namely, SC1/2 (seed coat, 3/15 days after flowering), An (anther, 1~3 days before flowering), SO (stigma and ovary, 1~3 days before flowering), ImSe (immature endosperm, 15 days after flowering), Pan5/10/20 (panicles harvested before heading at lengths of 5/10/20 cm), and Car1/3/5/7/10/15/20 (caryopses, 1/3/5/7/10/15/20 days after flowering) [23]. Three biological replicates were produced for every tissue and each biological replicate was collected and pooled together from over 30 plants. To ensure the accuracy of the results, three technical replicates were set for each biological replicate in quantitative real-time PCR (qRT-PCR).
2.2. Identification and Phylogenetic Analysis of TMT Genes
In this study, two approaches were adopted to identify TMT proteins in seven tested genomes: hidden Markov model (HMM) and BLAST homology search. Among them, the Bd, Hv, Si, Sb, Zm, and Or genomes were downloaded from Ensembl Plants release 41 (http://plants.ensembl.org/index.html, accessed on 1 December 2022) and the Os (MSU 7.0) genome was obtained from the TIGR database (http://rice.plantbiology.msu.edu, accessed on 1 December 2022) [24]. The HMM profile of the Sugar_tr (PF00083) was obtained from Pfam (http://pfam.xfam.org/, accessed on 1 December 2022) were used to search TMT protein sequences against seven tested crops’ protein sequence datasets with default parameters using HMMER 3.2.1. Then, all identified proteins from the HMM homology searches were researched through the blastP method by local ncbi-blast-2.7.1+. Finally, all candidate protein sequences were submitted to the SMART website (http://smart.embl-heidelberg.de/, accessed on 10 December 2022) and the Pfam website (http://pfam.xfam.org/search/sequence, accessed on 10 December 2022) to check the completeness of the Sugar_tr domain.
All identified TMT protein sequences were aligned by ClustalW using PAM protein weight matrix, and a phylogeny tree was produced via the MEGA 6.0 software using the maximum likelihood (ML) method with 1000 bootstrap replicates [1,14,24]. According to clustering results, all TMT genes were divided into different clades.
2.3. Microsynteny Analysis, Consevered Motifs, and Gene Structure
The collinearity gene pairs of Os with Bd, Hv, Or, Sb, Si, and Zm were analyzed by using the Multiple Collinearity Scan toolkit X version (MCScanX), as described previously [25], and visualized using the ”dual synteny plotter” in TBtools [26]. Twenty conserved motifs were set in the MEME program (http://meme-suite.org/tools/meme, accessed on 20 December 2022) with the following parameters: motif width between 6 and 100, and other default parameters [1]. Genomic structures of TMT genes were gained from genomic annotation files. Finally, we used TBtools to visualize the result of the phylogenetic tree, gene structures, and conserved motifs of TMT genes [26].
2.4. Quantitative Analysis of TMT Genes in Rice
All RNAs were extracted using TRIzol Ragent (Invitrogen, Beijing, China) and reversed to cDNAs using HiScript III 1st Strand cDNA Synthesis SuperMix for qPCR (Cat No. 11141ES60, Yeasen, Shanghai, China). The qRT-PCR reactions (10 μL) were formulated using Hieff UNICON Universal Blue qPCR SYBR Green Master Mix (Cat No. 11184ES08; Yeasen, Shanghai, China), following the manufacturer’s protocol. The primers were designed by Primer 5.0 (Table S1). All qRT-PCR reactions were conducted on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The actin gene was used as an internal control [14,23] and relative expression values were calculated by the 2−ΔΔCT method based on 3 biological replicates ×3 technical replicates [1,14]. All heatmaps were created using R package (pheatmap).
2.5. Expression Analysis of Rice TMT Genes under Cold and Salt Stress
Normalized intensities data of three-leaf-stage shoots and roots of TNG67 (indica) and TCN1 (japonica) under cold stress (4 °C, GSE57895, 96 microarray datasets) and salt stress (250 mM NaCl treatment, GSE76613, 96 microarray datasets) were download from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/, accessed on 20 December 2022). The expression changes in TMT genes after stress treatment were calculated using the formula: fold change (FC) = average expression amount of the treatment groups/average expression amount of the control groups.
2.6. Population Genetic Differentiation Coefficient (Fst, Fixation Index) of Rice TMT Genes
Based on the published rice 3K resequencing data (3K RG 1M GWAS SNP Dataset, all chromosomes, http://iric.irri.org/, accessed on 20 December 2022) including 1770 indica rice and 850 japonica rice, the Fst values of SNPs in the open reading frame (ORF) of the TMT genes identified in this study were calculated using the vcftools software. The average Fst value of all SNPs in the ORF of each gene was recorded as the Fst value of this gene.
3. Results
3.1. Identification of TMT Genes in Gramineae Crop Genomes
A total of 35 TMT genes were identified in seven Gramineae crop genomes, namely, six genes in Bd, three genes in Hv, six genes in Or, six genes in Os, four genes in Sb, six genes in Si, and four genes in Zm (Table 1). The identified TMT genes were named BdTMT, HvTMT, OrTMT, OsTMT, SbTMT, SiTMT, and ZmTMT followed by a number according to the chromosomal order in each genome, in accordance with the previous TMT study [27,28]. To understand the evolutionary relationships of these TMT genes among Gramineae crops, an ML phylogenetic tree was generated using MEGA 6.0. As shown in Table 2 and Figure 1, all TMT proteins were divided into three clades: I, II, and III. Clade III belonged to single-gene clade and had fewer TMT genes than those in clades I and II (Figure 1). Further analysis found that no clade III TMT gene existed in Hv. A selective pressure analysis showed that all three clades were under negative selective forces. We speculated that the number difference between different species may be caused by genetic duplication events. However, the MCScanX results showed that no gene duplication events were found in all tested species. These results indicated that the ancestors of Gramineous plants may have six TMT ancestor genes and that different gramineous species have unequal loss of TMT genes during species differentiation, which resulted in the current difference in the number of TMT genes.

Table 1.
The detailed information of tonoplast monosaccharide transporter (TMT) genes in seven Gramineae crops.

Table 2.
Gene numbers of TMT genes in seven tested species and Tajima’s values of the three clades.
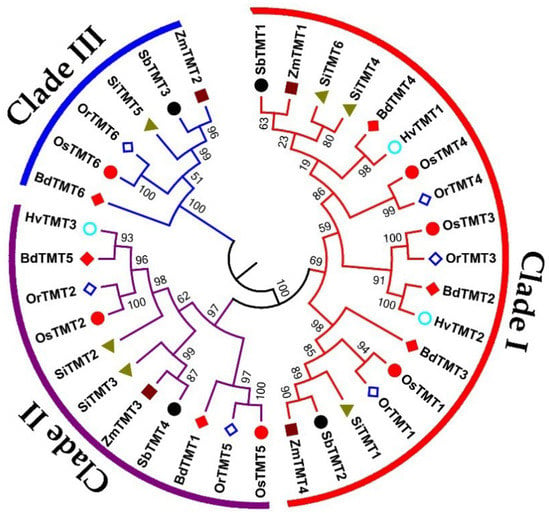
Figure 1.
A maximum likelihood (ML) phylogeny tree of TMT protein sequences from O. sativa ssp. japonica (Os), O. rufipogon (Or), H. vulgare (Hv), S. bicolor (Sb), B. distachyon (Bd), S. italica (Si), and Z. mays (Zm). The genes in the red, purple, and blue range belong to clades Ⅰ, Ⅱ, and Ⅲ, respectively.
3.2. Collinearity Gene Pairs, Intron/Exon Structure, and Conserved Motifs
The chromosome location results of TMT genes showed that the TMT genes were unevenly distributed on all chromosomes of the tested species. TMT genes were found only in certain segments of some chromosomes (Figure S1). All these segments had good linear relationships between the Os genome and other tested crop genomes (Figure 2). Here, we identified four, two, eight, four, five, and two collinearity gene pairs between Os and Bd, Hv, Or, Sb, Si, and Zm, respectively (Figure 2). There were more collinearity gene pairs between Os and Or, Si, Bd, and Sb than between OS and Hv and Zm, which basically supported the evolutionary distance of the tested species.
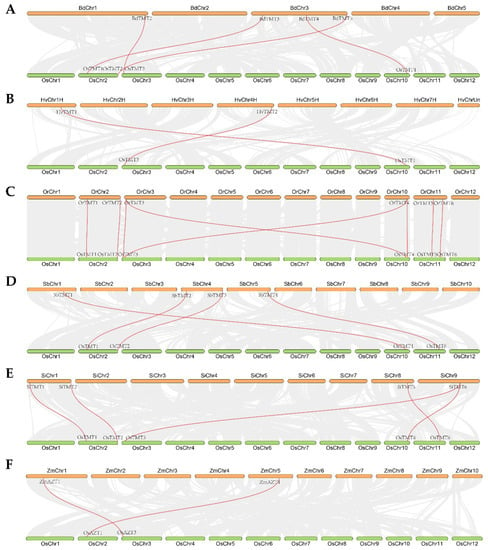
Figure 2.
Collinearity relationships of TMT genes between O. sativa ssp. japonica (Os) and B. distachyon (Bd), H. vulgare (Hv), O. rufipogon (Or), S. bicolor (Sb), S. italica (Si), and Z. mays (Zm), respectively: (A) Os vs. Bd; (B) Os vs. Hv; (C) Os vs. Or; (D) Os vs. Sb; (E) Os vs. Si; (F) Os vs. Zm.
In addition, intron/exon structure and conserved motifs of TMT genes in Gramineae crops revealed that TMT genes were very conserved in different species because they had similar structural features (Figure 3). Genes from the same clade shared similar gene structures and conserved motif organizations, while genes from different clades showed different gene structures and conserved motif organizations. For example, clades I (5–8 exons) and III (5–6 exons) possessed more exons than clade II (1–2 exons). In summary, TMT genes were relatively conserved among Gramineae crops, while structural differentiaition had occurred in different clades during the process of evolution, which provided a structural basis for the functional differentiation of TMT genes in species. Of course, this speculation needs to be further supported by evidence from gene expression data.
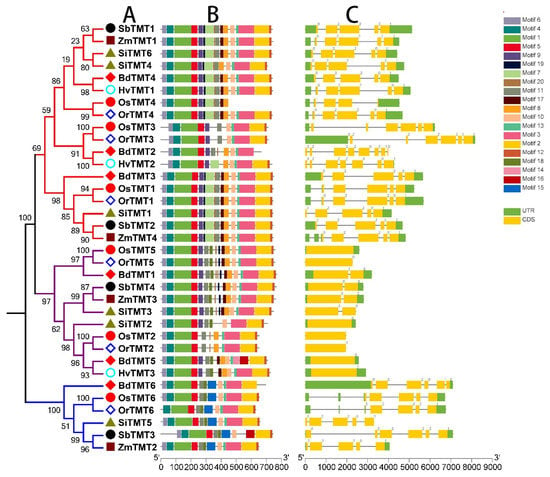
Figure 3.
Phylogenetic tree (A), motif compositions (B), and exon/intron structure (C) of the TMT genes in seven Gramineae crops. Different branches in the phylogenetic tree represent different clades. The relative lengths of proteins and genes can be estimated by using the gray bars. Untranslated regions (UTRs), exons, and introns are represented by green blue boxes, yellow boxes and gray lines, respectively.
3.3. Expression Patterns of TMT Genes in Various Tissues of Rice
Our expression profiling results revealed that rice TMT genes showed different expression levels in various tissues at different development stages or in 15 reproductive tissues (Figure 4). OsTMT1 and OsTMT4 showed higher expression levels in most tissues as compared with OsTMT3, OsTMT2, and OsTMT6 (Figure 4A). OsTMT5 showed specific expression of panicle, spike, and pollen (Figure 4A), suggesting that this gene may play roles in these specific tissues.
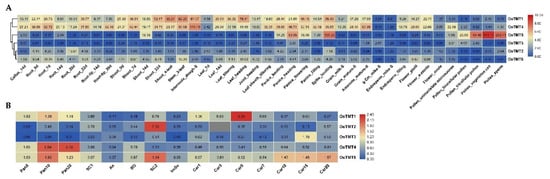
Figure 4.
(A,B) Expression profiles of rice TMT genes in different tissues; (B) SC1/2 (seed coat, 3/15 days after flowering), An (anther, 1~3 days before flowering), SO (stigma and ovary, 1~3 days before flowering), ImSe (immature endosperm, 15 days after flowering), Pan5/10/20 (panicles harvested before heading at lengths of 5/10/20 cm), and Car1/3/5/7/10/15/20 (caryopses, 1/3/5/7/10/15/20 days after flowering).
OsTMT1 showed the highest expression level in Car5. OsTMT2 was highly expressed in SC2. OsTMT3 had the highest expression level in Car15. However, OsTMT4 displayed the highest expression levels in pan10 and pan20. OsTMT6 showed relatively high expression levels in pan10 and SC2 (Figure 4B). These clearly differentiated expression profiles further prove that rice TMT genes had undergone functional differentiations.
3.4. Expression Patterns of TMT Genes in Rice under Cold or Salt Stress
It is well known that salt stress and cold stress are the two main abiotic stresses that affect the normal growth and yield of rice [29,30]. Indica and japonica, as two subspecies, show different levels of stress tolerance under the same strength of stress pressure [30]. In addition, the roles of rice TMT genes in salt stress and cold stress are still unclear.
In this study, the expression changes in rice TMT genes under cold and salt stresses were studies (Figure 5). We found that some TMT genes in shoots and roots had different transcriptional responses to salt and cold stress, such as OsTMT4 and OsTMT1 in shoots and roots under salt stress (Figure 5A), and OsTMT2 in shoots and roots under cold stress (Figure 5B). Under the same stress treatment, most of the TMT genes showed similar expression profiles in indica and japonica rice, but some genes were significantly different in indica and japonica rice, namely, OsTMT3 and OsTMT5 under salt stress (Figure 5A) and OsTMT3 under cold stress (Figure 5B). The expression of OsTMT6 was induced in both shoots and roots at 3 h and 24 h after salt stress treatment in two subspecies. Additionally, OsTMT3 showed different expression changes in indica and japonica rice after cold stress treatment. This gene was upregulated in the root of TNG67 at 3 h, 24 h, and Re: 24 h, while it was upregulated in the root of TCN1 at only 24 h (Figure 5B).
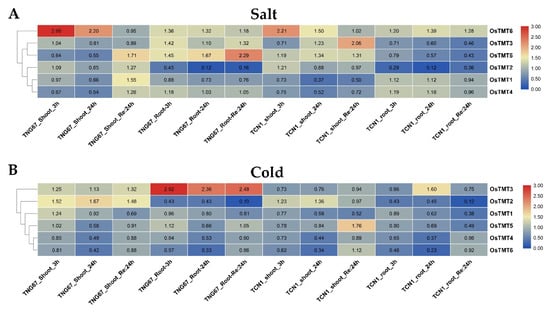
Figure 5.
Expression changes in the rice 3-leaf seedling under salt (A) and cold (B) stress. 3 h, 24 h, and Re: 24 h mean 3 h after treatment, 24 h after treatment, and recovering 24 h after termination of treatment.
3.5. Fst Values of Rice TMT Genes
Previous studies have reported that an Fst value of 0–0.05 indicates that there is no differentiation among the populations; an Fst value of 0.05–0.15 indicates that the populations are moderately differentiated; an Fst value of 0.15–0.25 indicates that the populations are highly differentiated; an Fst value greater than 0.25 indicates complete differentiation [31,32,33]. In this study, OsTMT2 was moderately differentiated between indica and japonica rice populations (0.0772). OsTMT6 was highly differentiated between indica and japonica rice populations (0.2476). OsTMT1, OsTMT3, OsTMT4, and OsTMT5 were completely differentiated between indica and japonica rice populations (>0.25) (Table 3). In particular, the Fst value of OsTMT4 was 0.9304, indicating that the gene was extremely differentiated between indica and japonica rice populations. These results indicated that the rice TMT gene family was under different selection pressures in the process of rice subspecies differentiation and later selection breeding.

Table 3.
Fst values of rice TMT genes.
4. Discussion
4.1. TMT Gene Lose in Gramineae Crop Genomes during the Process of Evolution
With the development of genome sequencing technology, more and more gene families have been identified in a large number of plants, such as GH3, GT8 and the AGC gene family [14,34,35,36]. Benefiting from genome-wide bioinformatics coupled with quantitative analysis, we identified and characterized the TMT gene family in Gramineae crops. Several studies have reported that gene duplication events play crucial roles in gene family expansion [5,14,37,38]. Our earlier study also found that duplication events led to the expansion of the rice STP, ERD, and PLT gene families (these all belong to the MST family) [28]. However, no gene duplication event of the TMT gene family was found in these seven species. On the contrary, gene lose may have occurred in some Gramineae crops. Clade III was lost in Hv. In addition, we found that there were no direct positive correlations between the number of TMT genes and plant genome sizes or whole genome duplication events. For example, the genome size of Hv is 4.79 Gbp with only three TMT genes, but the genome size of Os is 382.78 Mbp with six TMT genes. Swigoňová reported that Z. mays underwent one specific WGD more than other Gramineae plants [39]. However, in this study, Z. mays had fewer TMT genes than Bd, Or, Os, and Si. Taken together, the TMT gene tends to be genetically lost rather than expanded during the differentiation of gramineous species.
4.2. Functional Differentiation of Rice TMT Genes Involves Multiple Tissue Development Processes and Cold Stress Responses
Previous studies have shown that genes with the same function have similar gene structures and expression profiles [40,41,42]. To characterize the putative function of TMT genes, the gene structures and tissue expression profiles of TMT genes in rice were analyzed. As a result, the differential expression profiles of TMT genes indicated that the rice TMT genes had undergone functional differentiations. Expression profiling results revealed that rice TMT genes showed different expression levels in various tissues at different development stages or in 15 reproductive tissues (Figure 4). OsTMT1 and OsTMT4 showed higher expression levels in most tissues than OsTMT3, OsTMT2, and OsTMT6 (Figure 4A). In addition, some genes were highly expressed at a certain stage of rice organ development (e.g., OsTMT1 in Car7, OsTMT2 in SC2, and OsTMT3 in Car15), which demonstrated that these TMT genes are involved in rice reproductive development.
Salt and cold are the major abiotic stresses that frequently affect the growth, development, and food yield of crops in many countries [14,43,44]. The mechanism by which TMT genes promote sugar transportation or accumulation has been studied extensively [8,42,45], but it is not clear whether TMT genes play roles under abiotic stress. In this study, OsTMT3 showed obvious upregulation under salt stresses. In addition, we found that some TMT genes in shoots and roots had different transcriptional responses to salt and cold stress, such as OsTMT4 and OsTMT1 in shoots and roots under salt stress (Figure 5A) and OsTMT2 in shoots and roots under cold stress (Figure 5B). These results revealed that TMT genes are associated with salt stress and that the OsTMT3 gene could be a good candidate gene for rice resistance breeding under cold stress.
4.3. Different Selection Pressures May Promote the Expression Differentiations and Functional Differentiations of TMT Genes
In this study, the phylogenetic tree, gene structure, and protein motifs all supported the division of TMT genes into three clades. In addition, the expression patterns of the rice TMT genes from three clades were varied in various tissues or under abiotic stresses. The Tajima’s D values of the three clades were different, suggesting that the TMT genes of the three clades were under different intensities of selection pressure. It is worth noting that the expression profiles of some TMT genes in indica and japonica subspecies were also different. The Fst values of all TMT genes were greater than 0.05, and the Fst value of four TMT genes were greater than 0.25. These results suggested that TMT genes had been severely differentiated in the two indica and japonica populations, which may be a reason for the differential expressions of rice TMT genes in these two subspecies.
5. Conclusions
In the present study, a total of 35 TMT genes were identified in seven Gramineae crop genomes, and all TMT proteins could be subdivided into three clades. Subsequently, we carried out a systematic bioinformatics analysis at the genome-wide scale, including a phylogenetic analysis, exon/intron structures, conserved motifs, and collinearity relations. Our findings suggested that some TMT genes may have been lost in some Gramineae crops. Expression profiling of rice TMT genes revealed that rice TMT genes may act in multiple tissues and also respond to salt and cold stress. The comprehensive analysis of the TMT genes in this study will be useful for further research on biological functions and the evolution of TMT genes in Gramineae crops.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14061140/s1, Table S1: Primers of rice TMT genes used for qRT-PCR in this study, Figure S1: Chromosomal distribution of the TMT genes.
Author Contributions
M.Z. performed all of the experiments, analyzed the data, prepared the figures and tables, and wrote the paper; M.Z. and J.C. conceived and designed the experiments; X.D., Y.J. and G.Z. prepared parts of the figures and tables. All authors have read and agreed to the published version of the manuscript.
Funding
The work was funded by the ‘5511’ Collaborative Innovation project for High-quality Development and Surpasses of Agriculture between Government of Fujian Province and Chinese Academy of Agricultural Sciences (Grant no. XTCXGC2021002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Kong, W.; An, B.; Zhang, Y.; Yang, J.; Li, S.; Sun, T.; Li, Y. Sugar transporter proteins (STPs) in Gramineae crops: Comparative analysis, phylogeny, evolution, and expression profiling. Cells 2019, 8, 560. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-M.; Zheng, D.-M.; Li, L.-T.; Qiao, X.; Wei, S.-W.; Bai, B.; Zhang, S.-L.; Wu, J. Genome-wide function, evolutionary characterization and expression analysis of sugar transporter family genes in pear (Pyrus bretschneideri Rehd). Plant Cell Physiol. 2015, 56, 1721–1737. [Google Scholar] [CrossRef] [PubMed]
- Noiraud, N.; Maurousset, L.; Lemoine, R. Identification of a mannitol transporter, AgMaT1, in Celery Phloem . Plant Cell 2001, 13, 695–705. [Google Scholar] [CrossRef]
- Juchaux-Cachau, M.; Landouar-Arsivaud, L.; Pichaut, J.-P.; Campion, C.; Porcheron, B.; Jeauffre, J.; Noiraud-Romy, N.; Simoneau, P.; Maurousset, L.; Lemoine, R. Characterization of AgMaT2, a plasma membrane mannitol transporter from celery, expressed in phloem cells, including Phloem Parenchyma cells. Plant Physiol. 2007, 145, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Büttner, M. The monosaccharide transporter(-like) gene family in Arabidopsis . FEBS Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef]
- Kühn, C. A comparison of the sucrose transporter systems of different plant species. Plant Biol. 2003, 5, 215–232. [Google Scholar] [CrossRef]
- Kühn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 287–297. [Google Scholar] [CrossRef]
- Wormit, A.; Trentmann, O.; Feifer, I.; Lohr, C.; Tjaden, J.; Meyer, S.; Schmidt, U.; Martinoia, E.; Neuhaus, H.E. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 2006, 18, 3476–3490. [Google Scholar] [CrossRef]
- Cho, J.-I.; Burla, B.; Lee, D.-W.; Ryoo, N.; Hong, S.-K.; Kim, H.-B.; Eom, J.-S.; Choi, S.-B.; Cho, M.-H.; Bhoo, S.H. Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa). New Phytol. 2010, 186, 657–668. [Google Scholar] [CrossRef]
- Cheng, R.; Zhang, H.P.; Cheng, Y.S.; Wang, Y.Z.; Wang, G.M.; Zhang, S.L. In silico and expression analysis of the tonoplast monosaccharide transporter (TAT) gene family in Pyrus bretschneideri . J. Hortic. Sci. Biotech. 2018, 93, 366–376. [Google Scholar] [CrossRef]
- Schulze, W.X.; Schneider, T.; Starck, S.; Martinoia, E.; Trentmann, O. Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. Plant J. Cell Mol. Biol. 2012, 69, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Schulz, A.; Beyhl, D.; Marten, I.; Wormit, A.; Neuhaus, E.; Poschet, G.; Büttner, M.; Schneider, S.; Sauer, N.; Hedrich, R. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2 . Plant J. 2011, 68, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Wingenter, K.; Schulz, A.; Wormit, A.; Wic, S.; Trentmann, O.; Hoermiller, I.I.; Heyer, A.G.; Marten, I.; Hedrich, R.; Neuhaus, H.E. Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis . Plant Physiol. 2010, 154, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Gong, Z.; Zhong, H.; Zhang, Y.; Zhao, G.; Gautam, M.; Deng, X.; Liu, C.; Zhang, C.; Li, Y. Expansion and evolutionary patterns of glycosyltransferase family 8 in Gramineae crop genomes and their expression under salt and cold stresses in Oryza sativa ssp. japonica. Biomolecules 2019, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; Vogel, J.P.; Garvin, D.F. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 2010, 463, 763. [Google Scholar]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012, 30, 555. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Jacquemin, J.; Bhatia, D.; Singh, K.; Wing, R.A. The international Oryza map alignment project: Development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr. Opin. Plant Biol. 2013, 16, 147–156. [Google Scholar] [CrossRef]
- Dan, B.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Plant Bioinform. Methods Protoc. 2016, 1374, 115–140. [Google Scholar]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Jie, L.; Xiaolong, D.; Silan, C.; Chao, O.; Huiyun, S.; Jing, Y.; Yangsheng, L. Silencing of D-lactate dehydrogenase impedes glyoxalase system and leads to methylglyoxal accumulation and growth inhibition in rice. Front. Plant Sci. 2017, 8, 2071. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.-Q.; Wang, J.-L.; Li, S.-J. Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes 2019, 10, 401. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 10, 289660. [Google Scholar]
- Johnson, D.A.; Hill, J.P.; Thomas, M.A. The monosaccharide transporter gene family in land plants is ancient and shows differential subfamily expression and expansion across lineages. BMC Evol. Biol. 2006, 6, 64. [Google Scholar]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Kong, W.; Li, Y. A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef]
- Kong, W.; Zhong, H.; Gong, Z.; Fang, X.; Sun, T.; Deng, X.; Li, Y. Meta-analysis of salt stress transcriptome responses in different rice genotypes at the seedling stage. Plants 2019, 8, 64. [Google Scholar] [CrossRef]
- Li, R.; Jiang, T.; Xu, C.; Li, X.; Wang, X. Relationship between morphological and genetic differentiation in rice (Oryza sativa L.). Euphytica 2000, 114, 1–8. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.A.; Caballero, A. Characterization and conservation of genetic diversity in subdivided populations. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1367–1378. [Google Scholar] [CrossRef]
- Lin, F.; Ge, M.; Zhou, L.; Zhao, H. Genome-Wide Identification of Glyco-hydro-16 Family in Maize and Differentiation Analysis. Sci. Agric. Sin. 2016, 49, 2039–2048. [Google Scholar]
- Motorin, Y.; Marchand, V. Analysis of RNA Modifications by Second- and Third-Generation Deep Sequencing: 2020 Update. Genes 2021, 12, 278. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhang, Y.; Deng, X.; Li, S.; Zhang, C.; Li, Y. Comparative Genomic and Transcriptomic Analysis Suggests the Evolutionary Dynamic of GH3 Genes in Gramineae Crops. Front. Plant Sci. 2019, 10, 1297. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, X.; Zhou, M.; Yang, J.; Ke, S.; Li, Y. Genome-Wide Identification of the AGC Protein Kinase Gene Family Related to Photosynthesis in Rice (Oryza sativa). Int. J. Mol. Sci. 2022, 23, 12557. [Google Scholar] [CrossRef]
- Kong, W.; Zhong, H.; Deng, X.; Gautam, M.; Gong, Z.; Zhang, Y.; Zhao, G.; Liu, C.; Li, Y. Evolutionary analysis of GH3 genes in six Oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica . Plants 2019, 8, 30. [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E.J.M. Distribution and evolution of the lectin family in soybean (Glycine max). Molecules 2015, 20, 2868–2891. [Google Scholar] [CrossRef]
- Swigonova, Z. Close split of sorghum and maize genome progenitors. Genome Res. 2004, 14, 1916–1923. [Google Scholar] [CrossRef]
- Jacquemin, J.; Ammiraju, J.S.S.; Haberer, G.; Billheimer, D.D.; Yu, Y.; Liu, L.C.; Rivera, L.F. Fifteen million years of evolution in the Oryza genus shows extensive gene family expansion. Mol. Plant 2014, 7, 642–656. [Google Scholar] [CrossRef]
- Yan, H.B.; Pan, X.-X.; Jiang, H.-W.; Wu, G.-J. Comparison of the starch synthesis genes between maize and rice: Copies, chromosome location and expression divergence. Theor. Appl. Genet. 2009, 119, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.A.; Thomas, M.A. The monosaccharide transporter gene family in Arabidopsis and rice: A history of duplications, adaptive evolution, and functional divergence. Mol. Biol. Evol. 2007, 24, 2412–2423. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wensheng, W.; Fan, Z.; Jianli, D.; Zhikang, L.; Binying, F.; Jauhar, A. Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS ONE 2014, 9, e108020. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Cakir, B.; Giachino, R.R.A. VvTMT2 encodes a putative tonoplast monosaccharide transporter expressed during grape berry (Vitis vinifera cv. Sultanine) ripening. Plant Omics 2012, 5, 576–583. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).