An In Vitro Model of Glioma Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Purification and Culturing
2.2. Proteomic Studies
2.3. Statistics
2.4. Purification of EVs
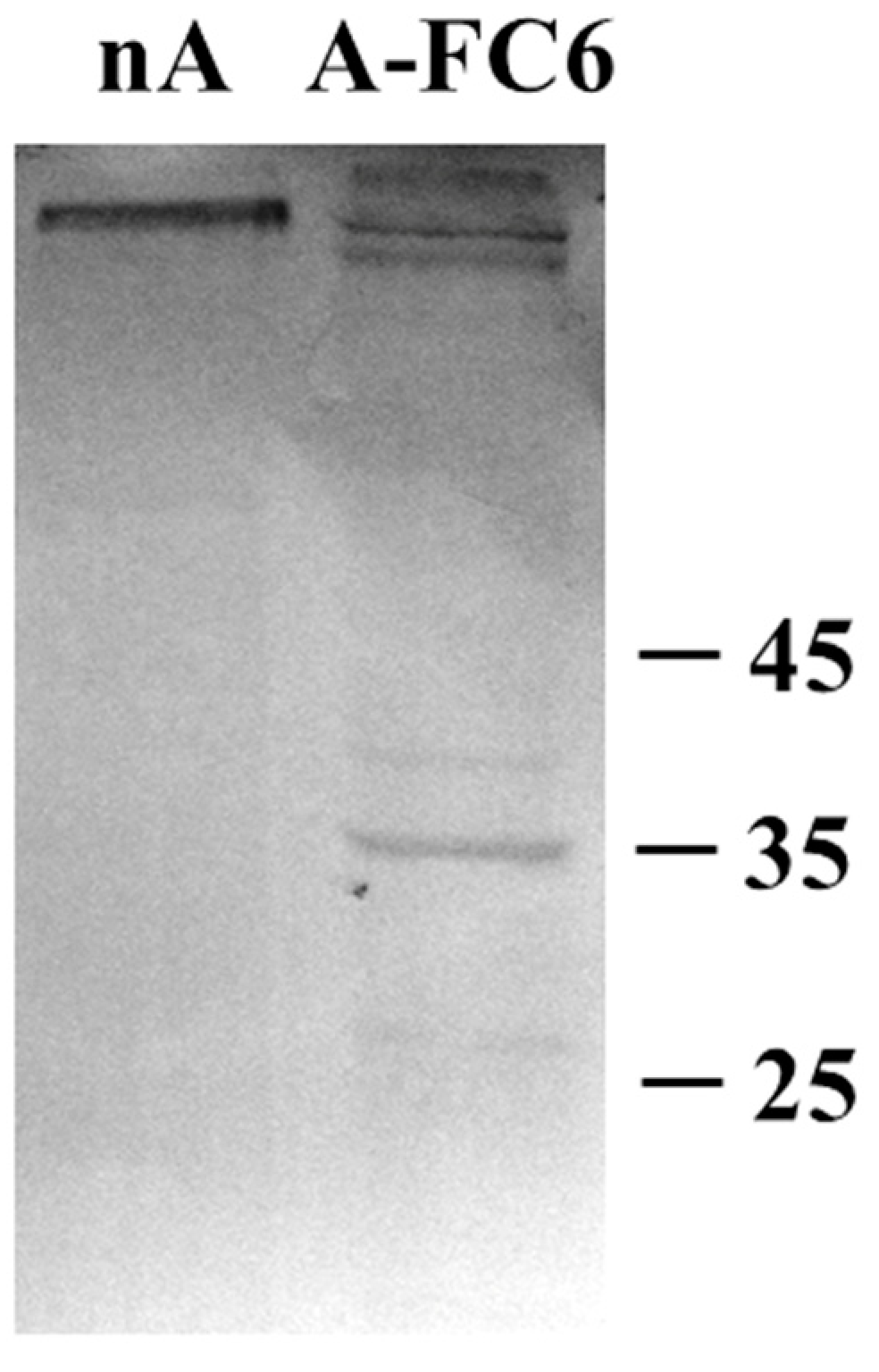
2.5. Western Blot Analysis
3. Results
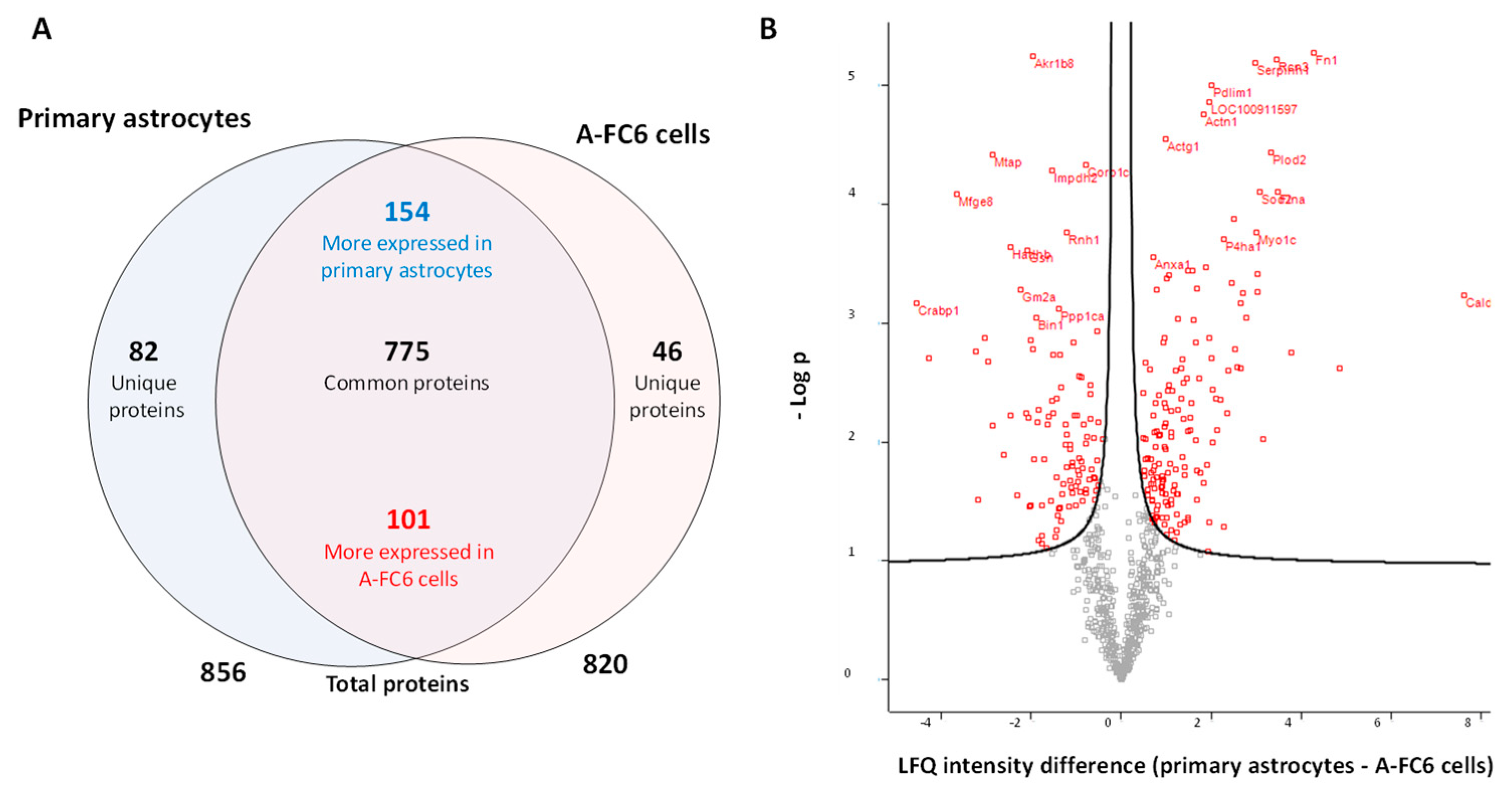
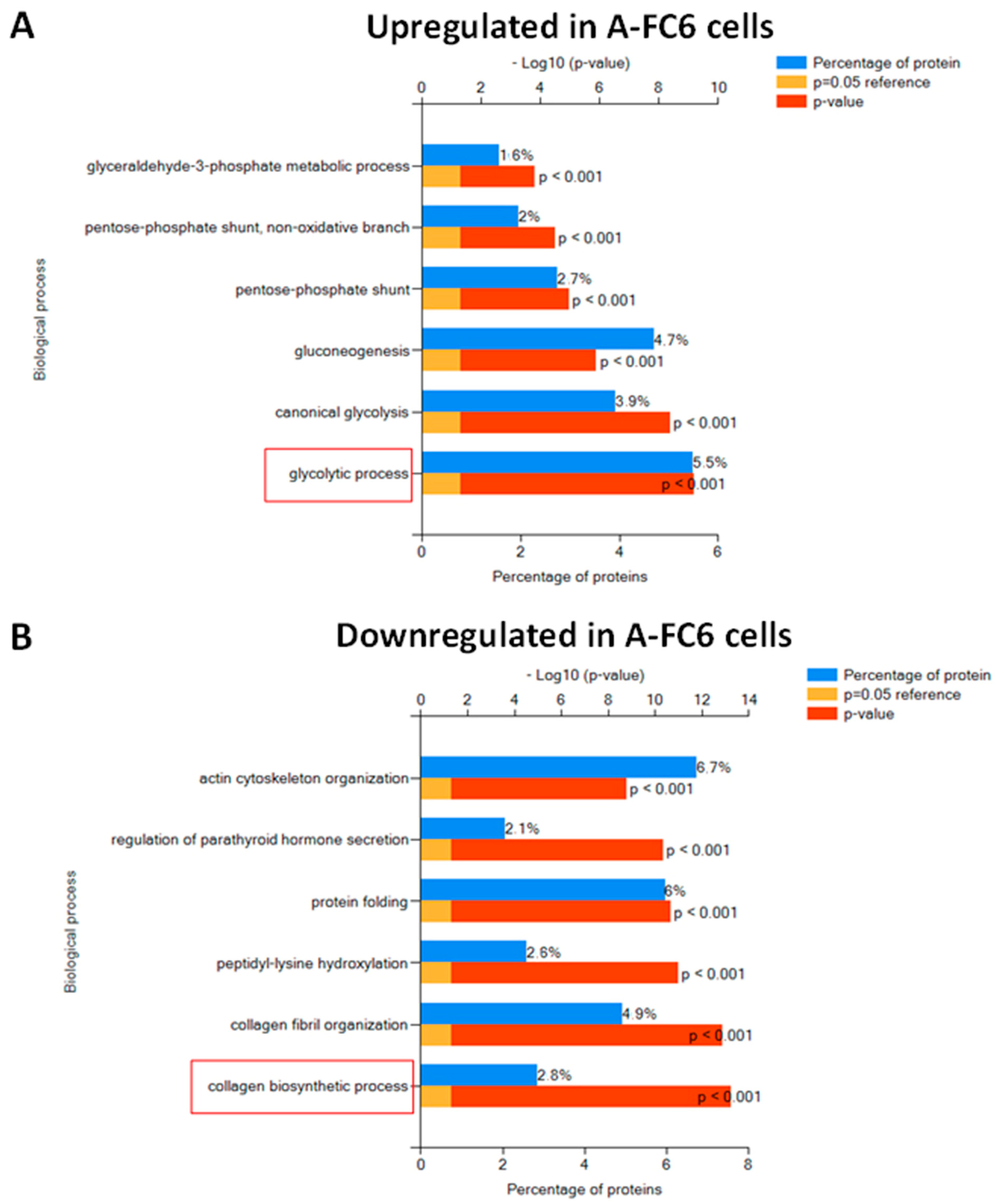
3.1. Comparative Proteomic Analysis
3.2. Comparative Proteomic Analysis with Respect to Chromosome #8 Gene Expression
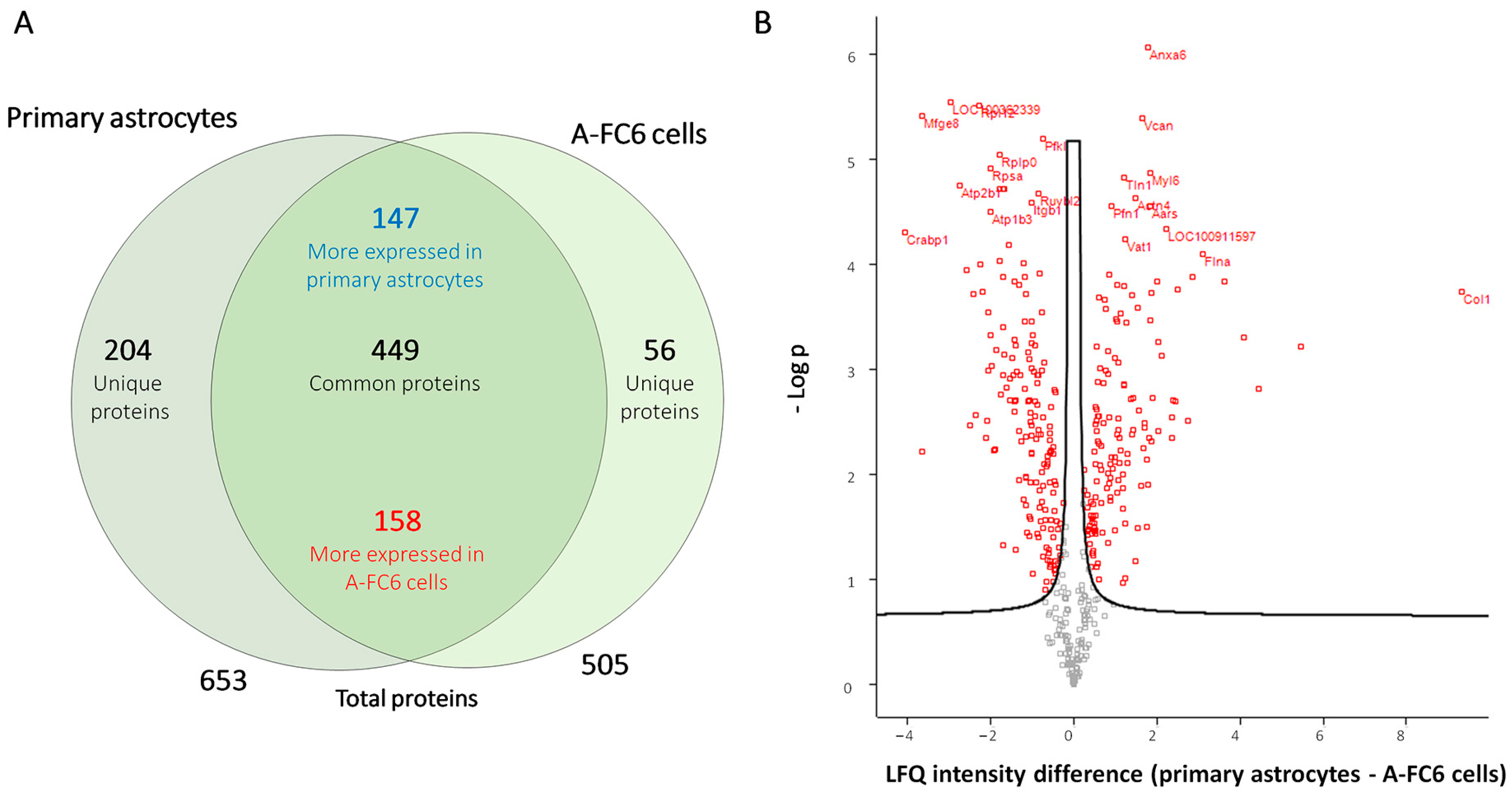
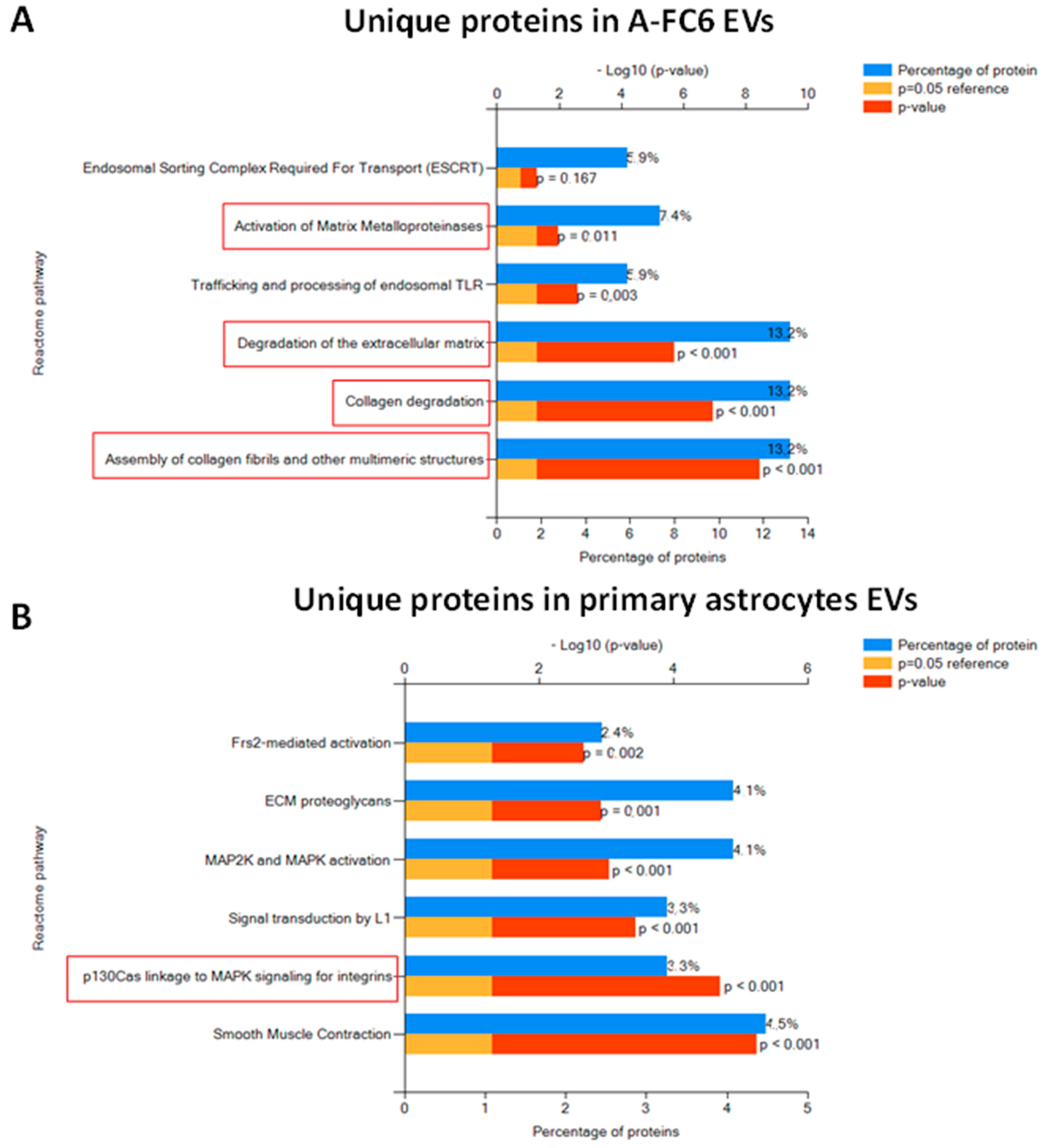
3.3. Comparative Proteomic Analysis of Extracellular Vesicles (EVs) Released from Either Normal or Transformed Astrocytes
4. Discussion
4.1. Proteomic Analysis
- The metalloproteinases Mmp3 and Mmp13, which are known to be involved in tumor invasion processes, since these proteases can degrade the extracellular matrix and also intracellular polymers, causing inflammation [34,35]. This suggests that the genomically more unstable line could favor a higher permeability of the brain parenchyma;
- Phospholipase C-delta1 (Pcld1), involved in cell cycle control [38];
- Poly (ADP-ribose) polymerase 3 (Parp3), involved in different transcriptional regulatory pathways in neuronal development [39].
- Von Willebrand factor (Vwa5a), a glycoprotein secreted by the endothelium and involved in some processes, such as platelet adhesion, development of the perivascular matrix and alteration of calcium signaling, glutamate absorption and the activity of some metalloproteinases [44];
- Archain 1 (Arcn1), which directs the maturation of the cerebellum and the intracellular vesicular traffic and seems to play a direct role also in neurodegenerative diseases [45];
- Inosine monophosphate dehydrogenase-2 (Impdh2), involved in the biosynthesis of GTP and in the support of cell proliferation [46];
4.2. EVs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Modrek, A.S.; Bayin, N.S.; Placantonakis, D.G. Brain stem cells as the cell of origin in glioma. World J. Stem Cells 2014, 26, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Prim. 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Pan, Y.; Gutmann, D.H. The power of the few. Genes Dev. 2017, 31, 1177–1179. [Google Scholar] [CrossRef]
- Schiera, G.; Di Liegro, C.M.; Di Liegro, I. Molecular Determinants of Malignant Brain Cancers: From Intracellular Alterations to Invasion Mediated by Extracellular Vesicles. Int. J. Mol. Sci. 2017, 18, 2774. [Google Scholar] [CrossRef]
- Sampetrean, O.; Saya, H. Modeling phenotypes of malignant gliomas. Cancer Sci. 2018, 109, 6–14. [Google Scholar] [CrossRef]
- Jovčevska, I. Next Generation Sequencing and Machine Learning Technologies Are Painting the Epigenetic Portrait of Glioblastoma. Front. Oncol. 2020, 10, 798. [Google Scholar] [CrossRef]
- Fuller, G.N.; Scheithauer, B.W. The 2007 Revised World Health Organization (WHO) Classification of Tumours of the Central Nervous System: Newly Codified Entities. Brain Pathol. 2007, 17, 304–307. [Google Scholar] [CrossRef]
- Perry, A.; Wesseling, P. Histologic classification of gliomas. Handb. Clin. Neurol. 2016, 134, 71–95. [Google Scholar] [CrossRef]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.W.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, andTERTPromoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef]
- Leeper, H.E.; Caron, A.A.; Decker, P.A.; Jenkins, R.B.; Lachance, D.H.; Giannini, C. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget 2015, 6, 30295–30305. [Google Scholar] [CrossRef]
- Bronisz, A.; Godlewski, J.; Chiocca, E.A. Extracellular Vesicles and MicroRNAs: Their Role in Tumorigenicity and Therapy for Brain Tumors. Cell. Mol. Neurobiol. 2016, 36, 361–376. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Onizuka, H.; Masui, K.; Komori, T. Diffuse gliomas to date and beyond 2016 WHO Classification of Tumours of the Central Nervous System. Int. J. Clin. Oncol. 2020, 25, 997–1003. [Google Scholar] [CrossRef]
- Knisely, J.; Schulder, M.; Touat, M.; Idbaih, A.; Sanson, M.; Buckner, J.C.; Chakravarti, A.; Curran, W.J., Jr. Radiation plus Chemotherapy in Low-Grade Glioma. N. Engl. J. Med. 2016, 375, 490–491. [Google Scholar] [CrossRef] [PubMed]
- Tipping, M.; Eickhoff, J.; Robins, H.I. Clinical outcomes in recurrent glioblastoma with bevacizumab therapy: An analysis of the literature. J. Clin. Neurosci. 2017, 44, 101–106. [Google Scholar] [CrossRef]
- Xu, W.; Li, T.; Gao, L.; Zheng, J.; Shao, A.; Zhang, J. Efficacy and safety of long-term therapy for high-grade glioma with temozolomide: A meta-analysis. Oncotarget 2017, 8, 51758–51765. [Google Scholar] [CrossRef] [PubMed]
- Thurin, E.; Nyström, P.W.; Smits, A.; Werlenius, K.; Bäck, A.; Liljegren, A.; Daxberg, E.-L.; Jakola, A.S. Proton therapy for low-grade gliomas in adults: A systematic review. Clin. Neurol. Neurosurg. 2018, 174, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Escobar, A.; Gutierrez, M.; Tejada, R. Letter to the editor regarding “Proton therapy for low-grade gliomas in adults: A systematic review”. Clin. Neurol. Neurosurg. 2020, 196, 105974. [Google Scholar] [CrossRef]
- Salvatore, V.; Teti, G.; Focaroli, S.; Mazzotti, M.C.; Mazzotti, A.; Falconi, M. The tumor microenvironment promotes cancer progression and cell migration. Oncotarget 2017, 8, 9608–9616. [Google Scholar] [CrossRef]
- Alexandru, O.; Horescu, C.; Sevastre, A.-S.; Cioc, C.; Baloi, C.; Oprita, A.; Dricu, A. Receptor tyrosine kinase targeting in glioblastoma: Performance, limitations and future approaches. Contemp. Oncol. 2020, 24, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Caradonna, F.; Schiera, G.; Di Liegro, C.M.; Vitale, V.; Cruciata, I.; Ferrara, T.; D’oca, P.; Mormino, R.; Rizzo, S.M.A.; Di Liegro, I. Establishment and Preliminary Characterization of Three Astrocytic Cells Lines Obtained from Primary Rat Astrocytes by Sub-Cloning. Genes 2020, 11, 1502. [Google Scholar] [CrossRef] [PubMed]
- Schiera, G.; Di Liegro, C.M.; Di Liegro, I. Extracellular Membrane Vesicles as Vehicles for Brain Cell-to-Cell Interactions in Physiological as well as Pathological Conditions. BioMed. Res. Int. 2015, 2015, 152926. [Google Scholar] [CrossRef] [PubMed]
- Carreca, A.P.; Pravatà, V.M.; D’apolito, D.; Bonelli, S.; Calligaris, M.; Monaca, E.; Müller, S.A.; Lichtenthaler, S.F.; Scilabra, S.D. Quantitative Proteomics Reveals Changes Induced by TIMP-3 on Cell Membrane Composition and Novel Metalloprotease Substrates. Int. J. Mol. Sci. 2021, 22, 2392. [Google Scholar] [CrossRef]
- Leloup, C.; Allard, C.; Carneiro, L.; Fioramonti, X.; Collins, S.; Pénicaud, L. Glucose and hypothalamic astrocytes: More than a fueling role? Neuroscience 2016, 323, 110–120. [Google Scholar] [CrossRef]
- Alberini, C.M.; Cruz, E.; Descalzi, G.; Bessières, B.; Gao, V. Astrocyte glycogen and lactate: New insights into learning and memory mechanisms. Glia 2017, 66, 1244–1262. [Google Scholar] [CrossRef]
- Bak, L.K.; Walls, A.B.; Schousboe, A.; Waagepetersen, H.S. Astrocytic glycogen metabolism in the healthy and diseased brain. J. Biol. Chem. 2018, 293, 7108–7116. [Google Scholar] [CrossRef]
- Brown, A.M.; Ransom, B.R. Astrocyte glycogen and brain energy metabolism. Glia 2007, 55, 1263–1271. [Google Scholar] [CrossRef]
- Valtcheva, S.; Venance, L. Control of Long-Term Plasticity by Glutamate Transporters. Front. Synaptic Neurosci. 2019, 11, 10. [Google Scholar] [CrossRef]
- Volterra, A.; Meldolesi, J. Astrocytes, from brain glue to communication elements: The revolution continues. Nat. Rev. Neurosci. 2005, 6, 626–640. [Google Scholar] [CrossRef]
- Parpura, V.; Zorec, R. Gliotransmission: Exocytotic release from astrocytes. Brain Res. Rev. 2010, 63, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Schiera, G.; Di Liegro, C.M.; Di Liegro, I. Cell-to-Cell Communication in Learning and Memory: From Neuro- and Glio-Transmission to Information Exchange Mediated by Extracellular Vesicles. Int. J. Mol. Sci. 2019, 21, 266. [Google Scholar] [CrossRef] [PubMed]
- Covelo, A.; Araque, A. Lateral regulation of synaptic transmission by astrocytes. Neuroscience 2016, 323, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Gobin, E.; Bagwell, K.; Wagner, J.; Mysona, D.; Sandirasegarane, S.; Smith, N.; Bai, S.; Sharma, A.; Schleifer, R.; She, J.-X. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 2019, 19, 581. [Google Scholar] [CrossRef]
- Bassiouni, W.; Ali, M.A.M.; Schulz, R. Multifunctional intracellular matrix metalloproteinases: Implications in disease. FEBS J. 2021, 288, 7162–7182. [Google Scholar] [CrossRef]
- De Vrij, F.M.; Bouwkamp, C.G.; Gunhanlar, N.; Shpak, G.; Lendemeijer, B.; Baghdadi, M.; Gopalakrishna, S.; Ghazvini, M.; Li, T.M.; Quadri, M.; et al. Candidate CSPG4 mutations and induced pluripotent stem cell modeling implicate oligodendrocyte progenitor cell dysfunction in familial schizophrenia. Mol. Psychiatry 2019, 24, 757–771. [Google Scholar] [CrossRef]
- Tamburini, E.; Dallatomasina, A.; Quartararo, J.; Cortelazzi, B.; Mangieri, D.; Lazzaretti, M.; Perris, R. Structural deciphering of the NG2/CSPG4 proteoglycan multifunctionality. FASEB J. 2018, 33, 3112–3128. [Google Scholar] [CrossRef]
- Stallings, J.D.; Zeng, Y.X.; Narvaez, F.; Rebecchi, M.J. Phospholipase C-δ1 Expression Is Linked to Proliferation, DNA Synthesis, and Cyclin E Levels. J. Biol. Chem. 2008, 283, 13992–14001. [Google Scholar] [CrossRef]
- Rodriguez-Vargas, J.M.; Nguekeu-Zebaze, L.; Dantzer, F. PARP3 comes to light as a prime target in cancer therapy. Cell Cycle 2019, 18, 1295–1301. [Google Scholar] [CrossRef]
- Kawabe, H.; Brose, N. The ubiquitin E3 ligase Nedd4-1 controls neurite development. Cell Cycle 2010, 9, 2477–2478. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara, M.; Nishiguchi, Y.; Fujiwara, R.; Kirita, T.; Kuniyasu, H. NEDD 4 binding protein 2-like 1 promotes cancer cell invasion in oral squamous cell carcinoma. Virchows Arch. 2016, 469, 163–172. [Google Scholar] [CrossRef]
- Du, S.-S.; Sun, X.; Cen, J.; Shi, J.-X.; An, M.-X.; Zhao, W.-D. Dynamin-2 mediates clathrin-dependent endocytosis for amyloid-β internalization in brain microvascular endothelial cells. Microvasc. Res. 2021, 138, 104219. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, P.M.; Gamby, C.; Wells, D.; Fallon, J.; Vallee, R.B. Dynamin Isoform-specific Interaction with the Shank/ProSAP Scaffolding Proteins of the Postsynaptic Density and Actin Cytoskeleton. J. Biol. Chem. 2001, 276, 48458–48465. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Akkouh, I.A.; Vandenberghe, M.; Osete, J.R.; Hughes, T.; Heine, V.; Smeland, O.B.; Glover, J.C.; Andreassen, O.A.; Djurovic, S. A human iPSC-astroglia neurodevelopmental model reveals divergent transcriptomic patterns in schizophrenia. Transl. Psychiatry 2021, 11, 554. [Google Scholar] [CrossRef]
- Xu, X.; Kedlaya, R.; Higuchi, H.; Ikeda, S.; Justice, M.J.; Setaluri, V.; Ikeda, A. Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration. PLoS Genet. 2010, 6, e1000956. [Google Scholar] [CrossRef] [PubMed]
- Kofuji, S.; Sasaki, A.T. GTP metabolic reprogramming by IMPDH2: Unlocking cancer cells’ fuelling mechanism. J. Biochem. 2020, 168, 319–328. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Persaud, S.D.; Nhieu, J.; Wei, L.-N. Cellular Retinoic Acid–Binding Protein 1 Modulates Stem Cell Proliferation to Affect Learning and Memory in Male Mice. Endocrinology 2017, 158, 3004–3014. [Google Scholar] [CrossRef]
- Notaras, M.; Lodhi, A.; Fang, H.; Greening, D.; Colak, D. The proteomic architecture of schizophrenia iPSC-derived cerebral organoids reveals alterations in GWAS and neuronal development factors. Transl. Psychiatry 2021, 11, 541. [Google Scholar] [CrossRef]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. Extracellular Vesicle-Associated RNA as a Carrier of Epigenetic Information. Genes 2017, 8, 240. [Google Scholar] [CrossRef]
- Abbott, N.J.; Patabendige, A.A.K.; Dolman, D.E.M.; Yusof, S.R.; Begley, D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Schiera, G.; Proia, P.; Alberti, C.; Mineo, M.; Savettieri, G.; Di Liegro, I. Neurons produce FGF2 and VEGF and secrete them at least in part by shedding extracellular vesicles. J. Cell Mol. Med. 2007, 11, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
- Proia, P.; Schiera, G.; Mineo, M.; Ingrassia, A.M.R.; Santoro, G.; Savettieri, G.; Di Liegro, I. Astrocytes shed extracellular vesicles that contain fibroblast growth factor-2 and vascular endothelial growth factor. Int. J. Mol. Med. 2008, 21, 63–67. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schiera, G.; Cancemi, P.; Di Liegro, C.M.; Naselli, F.; Volpes, S.; Cruciata, I.; Cardinale, P.S.; Vaglica, F.; Calligaris, M.; Carreca, A.P.; et al. An In Vitro Model of Glioma Development. Genes 2023, 14, 990. https://doi.org/10.3390/genes14050990
Schiera G, Cancemi P, Di Liegro CM, Naselli F, Volpes S, Cruciata I, Cardinale PS, Vaglica F, Calligaris M, Carreca AP, et al. An In Vitro Model of Glioma Development. Genes. 2023; 14(5):990. https://doi.org/10.3390/genes14050990
Chicago/Turabian StyleSchiera, Gabriella, Patrizia Cancemi, Carlo Maria Di Liegro, Flores Naselli, Sara Volpes, Ilenia Cruciata, Paola Sofia Cardinale, Fabiola Vaglica, Matteo Calligaris, Anna Paola Carreca, and et al. 2023. "An In Vitro Model of Glioma Development" Genes 14, no. 5: 990. https://doi.org/10.3390/genes14050990
APA StyleSchiera, G., Cancemi, P., Di Liegro, C. M., Naselli, F., Volpes, S., Cruciata, I., Cardinale, P. S., Vaglica, F., Calligaris, M., Carreca, A. P., Chiarelli, R., Scilabra, S. D., Leone, O., Caradonna, F., & Di Liegro, I. (2023). An In Vitro Model of Glioma Development. Genes, 14(5), 990. https://doi.org/10.3390/genes14050990









