Evolutionary Adaptation of Genes Involved in Galactose Derivatives Metabolism in Oil-Tea Specialized Andrena Species
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peters, R.S.; Krogmann, L.; Mayer, C.; Donath, A.; Gunkel, S.; Meusemann, K.; Kozlov, A.; Podsiadlowski, L.; Petersen, M.; Lanfear, R.; et al. Evolutionary history of the Hymenoptera. Curr. Biol. 2017, 27, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Michener, C.D. The Bees of the World, 2nd ed.; The Johns Hopkins University Press: Baltimore, MD, USA, 2007. [Google Scholar]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Cane, J.H.; Sipes, S. Characterizing floral specialization by bees: Analytical methods and revised lexicon for oligolecty. In Plant-Pollinator Interactions: From Specialization to Generalization; Waser, N.M., Ollerton, J., Eds.; The University of Chicago Press: Chicago, IL, USA, 2006; pp. 99–121. [Google Scholar]
- Goulson, D. Bumblebees: Behaviour, Ecology, and Conservation; Oxford University Press Inc.: New York, NY, USA, 2010. [Google Scholar]
- Minckley, R.L.; Roulston, T.H. Incidental mutualisms and pollen specialization among bees. In Plant-Pollinator Interactions: From Specialization to Generalization; Waser, N.M., Ollerton, J., Eds.; The University of Chicago Press: Chicago, IL, USA, 2006; pp. 69–98. [Google Scholar]
- Bossert, S.; Wood, T.J.; Patiny, S.; Michez, D.; Almeida, E.A.B.; Minckley, R.L.; Packer, L.; Neff, J.L.; Copeland, R.S.; Straka, J.; et al. Phylogeny, biogeography and diversification of the mining bee family Andrenidae. Syst. Entomol. 2022, 47, 283–302. [Google Scholar] [CrossRef]
- Larkin, L.L.; Neff, J.L.; Simpson, B.B. The evolution of pollen diet: Host choice and diet breadth of Andrena bees (Hymenoptera: Andrenidae). Apidologie 2008, 39, 133–145. [Google Scholar] [CrossRef]
- Wood, T.J.; Roberts, S.P.M. An assessment of historical and contemporary diet breadth in polylectic Andrena bee species. Biol. Conserv. 2017, 215, 72–80. [Google Scholar] [CrossRef]
- Luan, F.; Zeng, J.; Yang, Y.; He, X.; Wang, B.; Gao, Y.; Zeng, N. Recent advances in Camellia oleifera Abel: A review of nutritional constituents, biofunctional properties, and potential industrial applications. J. Funct. Food. 2020, 75, 104242. [Google Scholar] [CrossRef]
- Wang, X.N. Research on Phenology and Blossom Biology of Oil-Tea Camellia. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2011. [Google Scholar]
- Huang, D.Y.; Ding, L.; Zhang, Y.Z.; Huang, H.R.; Yu, J.F.; Hao, J.S.; Zhu, C.D. Life history and relevant biological features of Andrena camellia Wu (Hymenoptera: Andrenidae). Acta Entomol. Sin. 2008, 51, 778–783. [Google Scholar]
- Xie, Z.; Chen, X.; Qiu, J. Reproductive failure of Camellia oleifera in the plateau region of China due to a shortage of legitimate pollinators. Int. J. Agric. Biol. 2013, 15, 458–464. [Google Scholar]
- Zhao, S.W. Management measure for honey colony in flowering period of Camellia oleifera. Apicult. China 1993, 5, 19–20. [Google Scholar]
- Ding, L.; Huang, D.Y.; Zhang, Y.Z.; Huang, H.R.; Li, J.; Zhu, C.D. Observation on the nesting biology of Andrena camellia Wu (Hymenoptera: Andrenidae). Acta Entomol. Sin. 2007, 50, 1077–1082. [Google Scholar]
- He, B.; Su, T.J.; Niu, Z.Q.; Zhou, Z.Y.; Gu, Z.Y.; Huang, D.Y. Characterization of mitochondrial genomes of three Andrena bees (Apoidea: Andrenidae) and insights into the phylogenetics. Int. J. Biol. Macromol. 2019, 127, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Su, T.J.; He, B.; Zhao, F.; Jiang, K.; Lin, G.; Huang, Z. Population genomics and phylogeography of Colletes gigas, a wild bee specialized on winter flowering plants. Ecol. Evol. 2022, 12, e8863. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.R. The pollinating bees on Camellia olifera with descriptions of 4 new species of the genus Andrena. Acta Entomol. Sin. 1977, 20, 199–204. [Google Scholar]
- Li, H.Y.; Luo, A.C.; Hao, Y.J.; Dou, F.Y.; Kou, R.M.; Orr, M.C.; Zhu, C.D.; Huang, D.Y. Comparison of the pollination efficiency of Apis cerana with wild bees in oil-seed camellia fields. Basic Appl. Ecol. 2021, 56, 250–258. [Google Scholar] [CrossRef]
- Kang, X.D.; Fan, Z.Y. Toxic contents of nectar of oil-tea flowers to honey bees. J. Bee 1991, 1, 8–10. [Google Scholar]
- Li, Z.; Huang, Q.; Zheng, Y.; Zhang, Y.; Li, X.; Zhong, S.; Zeng, Z. Identification of the toxic compounds in Camellia oleifera honey and pollen to honey bees (Apis mellifera). J. Agric. Food Chem. 2022, 70, 13176–13185. [Google Scholar] [CrossRef]
- Holden, H.M.; Rayment, I.; Thoden, J.B. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 2003, 278, 43885–43888. [Google Scholar] [CrossRef]
- Vinson, C.C.; Mota, A.P.Z.; Porto, B.N.; Oliveira, T.N.; Sampaio, I.; Lacerda, A.L.; Danchin, E.G.J.; Guimaraes, P.M.; Williams, T.C.R.; Brasileiro, A.C.M. Characterization of raffinose metabolism genes uncovers a wild Arachis galactinol synthase conferring tolerance to abiotic stresses. Sci. Rep. 2020, 10, 15258. [Google Scholar] [CrossRef]
- Elango, D.; Rajendran, K.; Van der Laan, L.; Sebastiar, S.; Raigne, J.; Thaiparambil, N.A.; El Haddad, N.; Raja, B.; Wang, W.; Ferela, A.; et al. Raffinose family oligosaccharides: Friend or foe for human and plant health? Front. Plant Sci. 2022, 13, 829118. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Slater, G.S.C.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinf. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, W.S.W.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Z.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Lomsadze, A.; Borodovsky, M. Identification of protein coding regions in RNA transcripts. Nucl. Acid. Res. 2015, 43, e78. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Kucukural, A.; Yukselen, O.; Ozata, D.M.; Moore, M.J.; Garber, M. DEBrowser: Interactive differential expression analysis and visualization tool for count data. BMC Genom. 2019, 20, 6. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q File manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Wen, Y.; Su, S.C.; Ma, L.Y.; Yang, S.Y.; Wang, Y.W.; Wang, X.N. Effects of canopy microclimate on fruit yield and quality of Camellia oleifera. Sci. Horticult. 2018, 235, 132–141. [Google Scholar]
- Feng, J.L.; Jiang, Y.; Yang, Z.J.; Chen, S.P.; El-Kassaby, Y.A.; Chen, H. Marker-assisted selection in C. oleifera hybrid population. Silvae Genet. 2020, 69, 63–72. [Google Scholar] [CrossRef]
- Deng, Y.; Yu, X.; Liu, Y. The role of native bees on the reproductive success of Camellia oleifera in Hunan Province, Central South China. Acta Ecol. Sin. 2010, 30, 4427–4436. [Google Scholar]
- Huang, D.; Su, T.; Qu, L.; Wu, Y.; Gu, P.; He, B.; Xu, X.; Zhu, C. The complete mitochondrial genome of the Colletes gigas (Hymenoptera: Colletidae: Colletinae). Mit. DNA Part A 2016, 27, 3878–3879. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Y.; He, B.; Gu, P.; Su, T.J.; Zhu, C.D. Discussion on current situation and research direction of pollination insects of Camellia oleifera. J. Environ. Entomol. 2017, 39, 213–220. [Google Scholar]
- Zhou, Q.S.; Luo, A.; Zhang, F.; Niu, Z.Q.; Wu, Q.T.; Xiong, M.; Orr, M.C.; Zhu, C.D. The first draft genome of the plasterer bee Colletes gigas (Hymenoptera: Colletidae: Colletes). Genome Biol. Evol. 2020, 12, 860–866. [Google Scholar] [CrossRef]
- Michalski, J.C.; Klein, A. Glycoprotein lysosomal storage disorders: K- and L-mannosidosis, fucosidosis and K-N-acetylgalactosaminidase deficiency. Biochim. Biophys. Acta 1999, 1455, 69–84. [Google Scholar] [CrossRef]
- Vallender, E.J.; Lahn, B.T. Positive selection on the human genome. Hum. Mol. Genet. 2004, 13, R245–R254. [Google Scholar] [CrossRef]
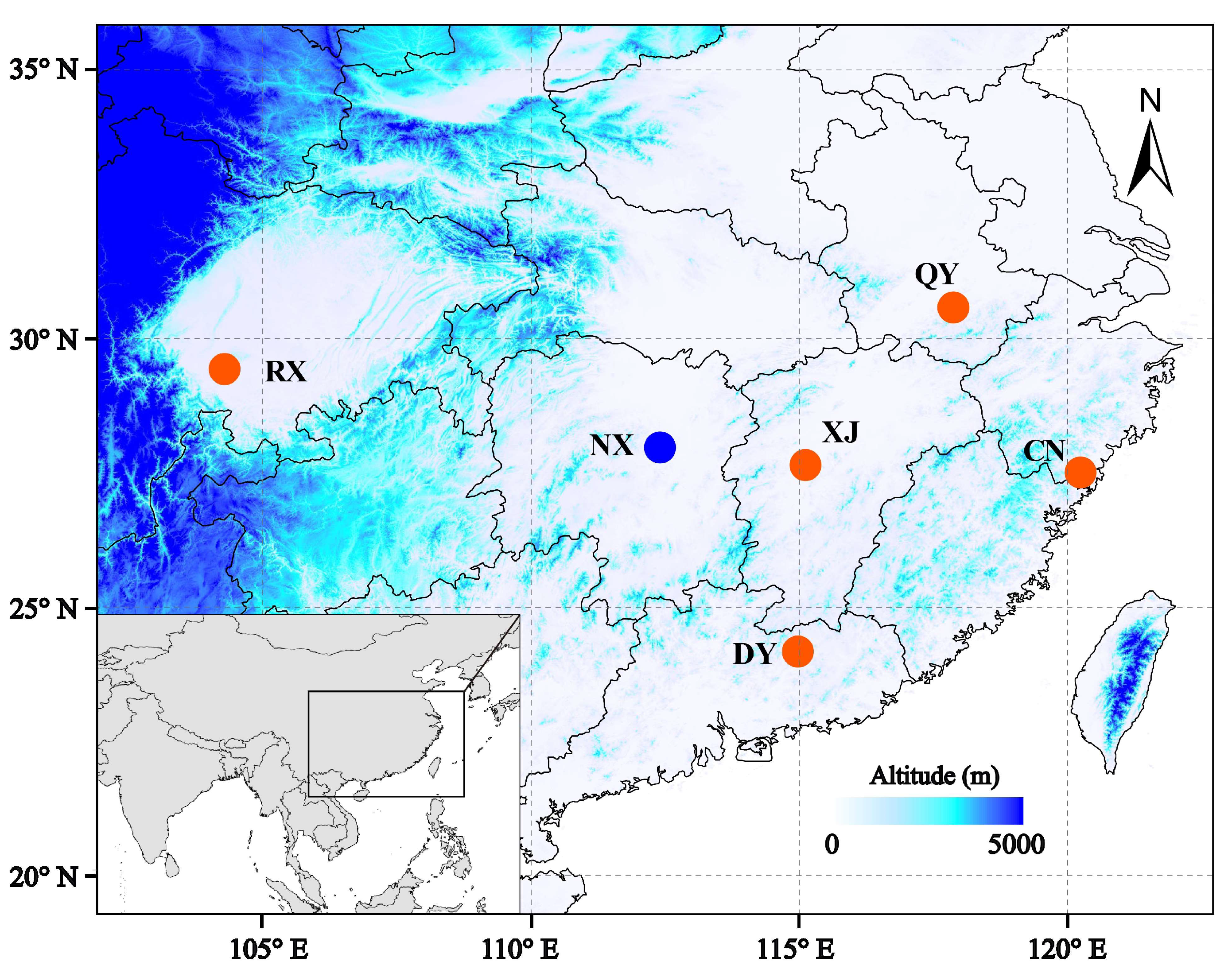
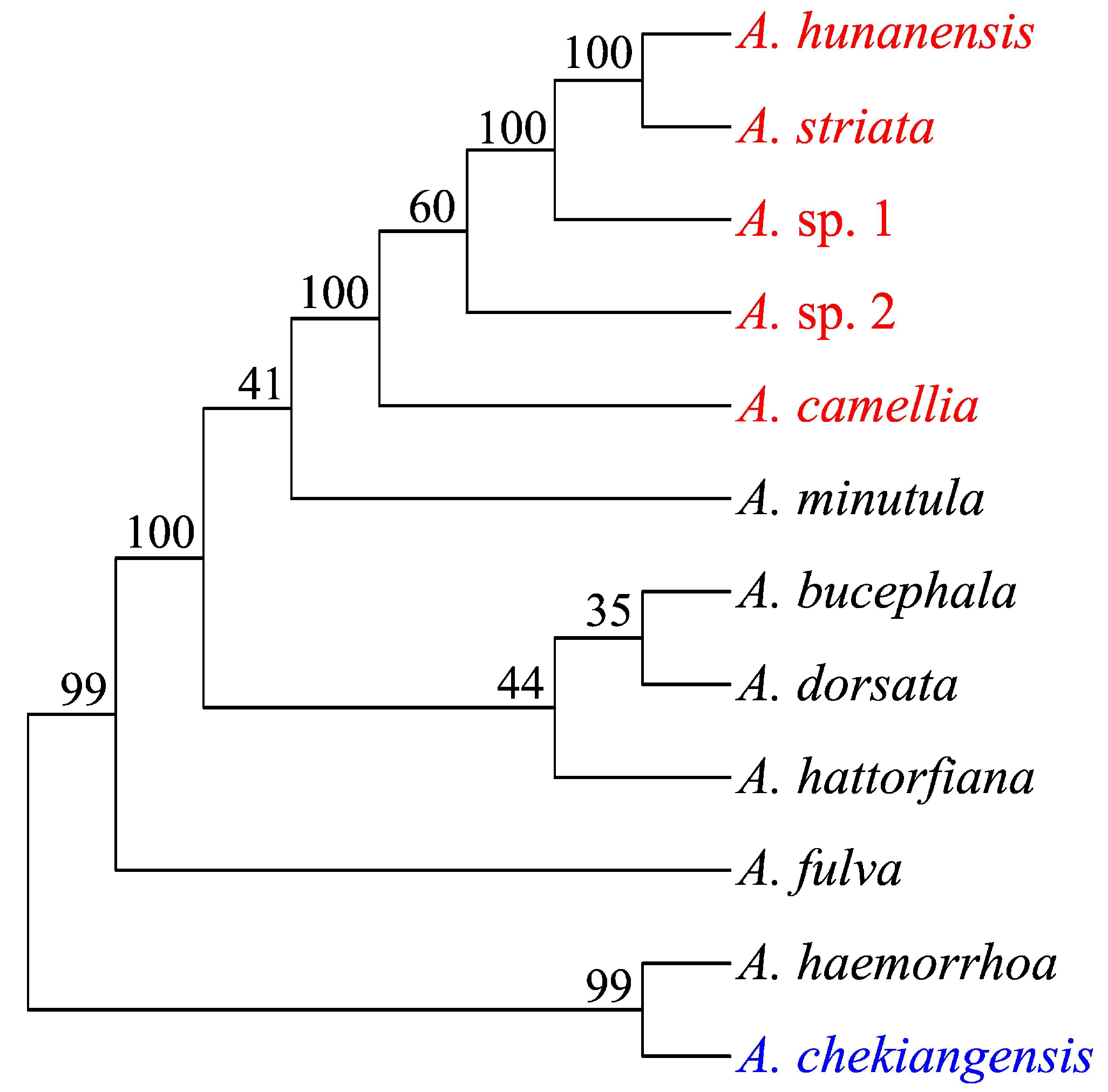
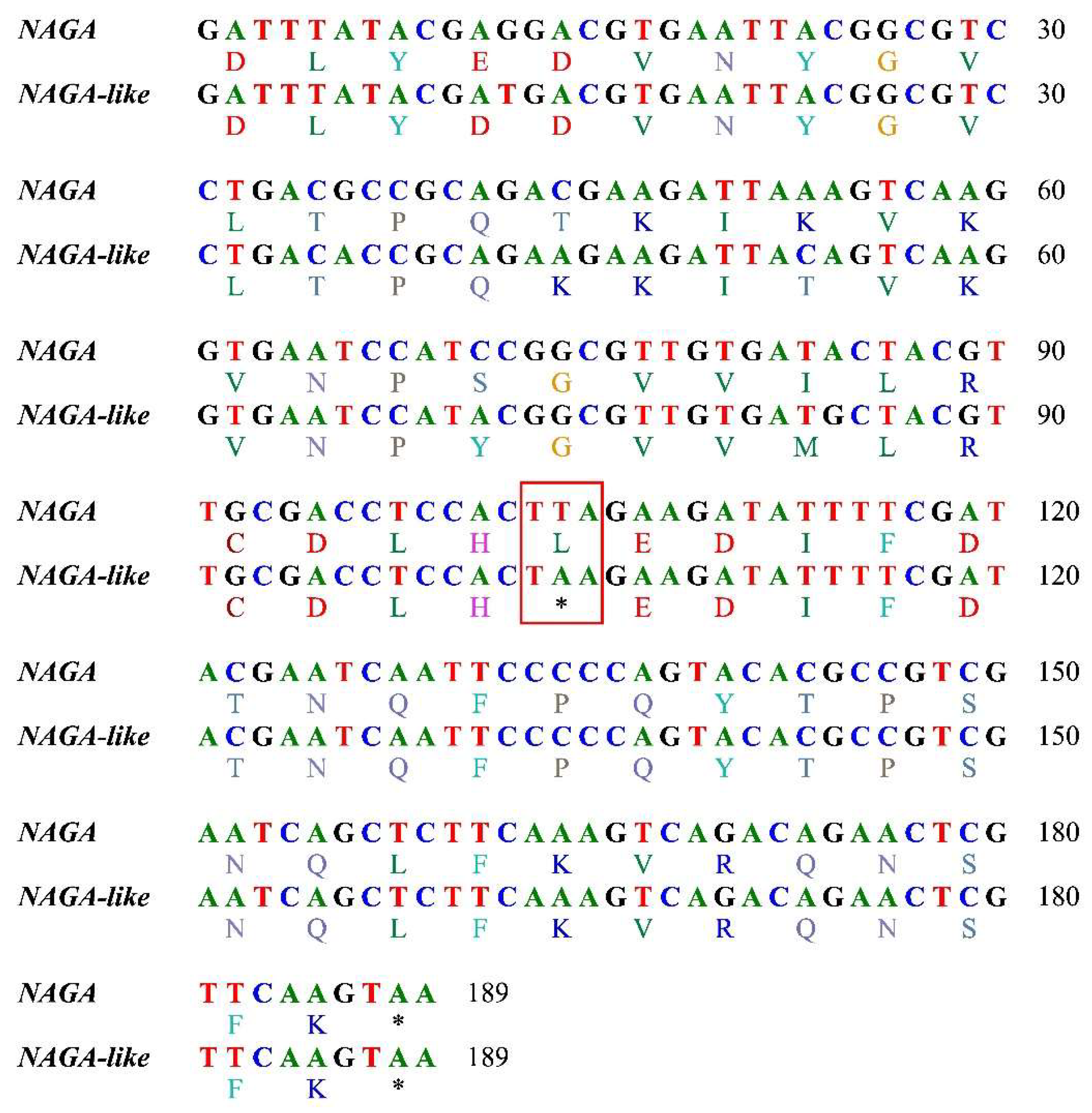

| Sample | Species | Location | Longitude | Latitude |
|---|---|---|---|---|
| XJ01 | A. camellia | Xiajiang, Jiangxi | 115.1285 | 27.6546 |
| QY01 | A. hunanensis | Qingyang, Anhui | 117.8796 | 30.5977 |
| RX01 | A. striata | Rongxian, Sichuan | 104.2913 | 29.4377 |
| CN02 | A. sp. 1 | Cangnan, Zhejiang | 120.2556 | 27.4591 |
| DY03 | A. sp. 2 | Dongyuan, Guangdong | 114.9792 | 24.1905 |
| NX04 | A. chekiangensis | Ningxiang, Hunan | 112.4206 | 27.9832 |
| Species | Reads | Assembly | ||
|---|---|---|---|---|
| Length (Gb) | Accession | Length (Mb) | N50 (Kb) | |
| A. camellia | 10.38 | SRR23869504 | 369.7 | 11.1 |
| A. hunanensis | 11.01 | SRR23869503 | 384.3 | 8.2 |
| A. striata | 9.79 | SRR23869502 | 393.1 | 8.5 |
| A. sp. 1 | 10.75 | SRR23869501 | 363.2 | 9.6 |
| A. sp. 2 | 9.74 | SRR23869500 | 353.5 | 14.7 |
| A. chekiangensis | 10.02 | SRR23869499 | 365.7 | 11.6 |
| Gene | Length | Variable Sites | Percent of Variable Sites |
|---|---|---|---|
| NAGA | 1320 | 16 | 1.21 |
| NAGA-like | 1239 | 33 | 2.66 |
| galM | 1077 | 29 | 2.69 |
| galK | 1182 | 24 | 2.03 |
| galT | 1152 | 15 | 1.30 |
| galE | 1098 | 8 | 0.73 |
| Gene | Foreground | Background | 2ΔlnL | p (df = 1) |
|---|---|---|---|---|
| NAGA | 0.049 | 0.023 | 1.412 | 0.234 |
| NAGA-like | 0.680 | 0.021 | 145.673 | <1.000 × 10−10 |
| galM | 0.251 | 0.360 | 1.283 | 0.257 |
| galK | 0.864 | 0.161 | 24.279 | 8.336 × 10−7 |
| galT | 0.387 | 0.088 | 12.761 | 3.540 × 10−4 |
| galE | 0.129 | 0.063 | 0.913 | 0.339 |
| Sample | Accession | Length (Gb) | Q30 (%) | GC (%) |
|---|---|---|---|---|
| Acam1 | SRR8335252 | 7.54 | 91.55 | 45.84 |
| Acam2 | SRR8335251 | 9.50 | 92.41 | 46.35 |
| Acam3 | SRR8335254 | 7.64 | 91.76 | 46.23 |
| Acam4 | SRR8335253 | 9.04 | 92.35 | 45.97 |
| Ache1 | SRR23869498 | 9.03 | 95.82 | 41.95 |
| Ache2 | SRR23869497 | 8.88 | 96.10 | 42.46 |
| Ache3 | SRR23869496 | 7.82 | 96.16 | 41.61 |
| Ache4 | SRR23869495 | 9.36 | 95.81 | 43.69 |
| Gene | TPM (Mean ± SD) | FC | Padj | |
|---|---|---|---|---|
| A. camellia | A. chekiangensis | |||
| NAGA | 574.88 ± 162.33 | 740.45 ± 470.32 | 0.639 | 0.387 |
| NAGA-like | 69,465.93 ± 7690.83 | 0 | +∞ | −∞ |
| galM | 537.64 ± 78.69 | 210.82 ± 211.28 | 2.14 | 0.118 |
| galK | 172.87 ± 47.92 | 41.77 ± 34.05 | 3.35 | 0.016 |
| galT | 987.77 ± 139.04 | 318.55 ± 57.03 | 2.99 | 1.781 × 10−12 |
| galE | 32.45 ± 10.07 | 21.82 ± 14.73 | 1.23 | 0.624 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, G.; Huang, Z.; He, B.; Jiang, K.; Su, T.; Zhao, F. Evolutionary Adaptation of Genes Involved in Galactose Derivatives Metabolism in Oil-Tea Specialized Andrena Species. Genes 2023, 14, 1117. https://doi.org/10.3390/genes14051117
Lin G, Huang Z, He B, Jiang K, Su T, Zhao F. Evolutionary Adaptation of Genes Involved in Galactose Derivatives Metabolism in Oil-Tea Specialized Andrena Species. Genes. 2023; 14(5):1117. https://doi.org/10.3390/genes14051117
Chicago/Turabian StyleLin, Gonghua, Zuhao Huang, Bo He, Kai Jiang, Tianjuan Su, and Fang Zhao. 2023. "Evolutionary Adaptation of Genes Involved in Galactose Derivatives Metabolism in Oil-Tea Specialized Andrena Species" Genes 14, no. 5: 1117. https://doi.org/10.3390/genes14051117
APA StyleLin, G., Huang, Z., He, B., Jiang, K., Su, T., & Zhao, F. (2023). Evolutionary Adaptation of Genes Involved in Galactose Derivatives Metabolism in Oil-Tea Specialized Andrena Species. Genes, 14(5), 1117. https://doi.org/10.3390/genes14051117






