Melatonin Protects the Apoptosis of Sheep Granulosa Cells by Suppressing Oxidative Stress via MAP3K8 and FOS Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Isolation and Culture of Granulosa Cells
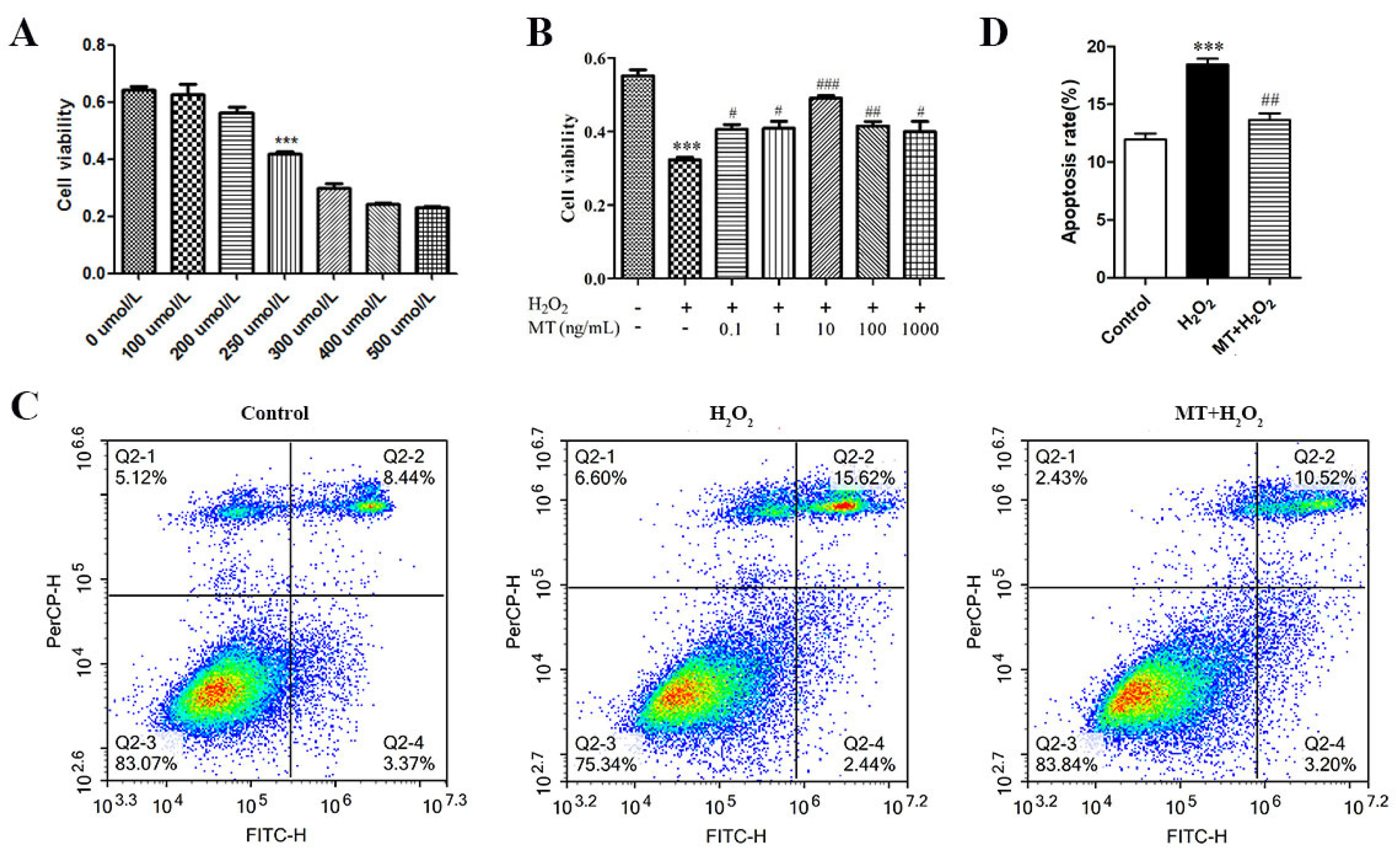
2.3. Cell Viability Assay
2.4. Flow Cytometric Analysis of Apoptotic cells
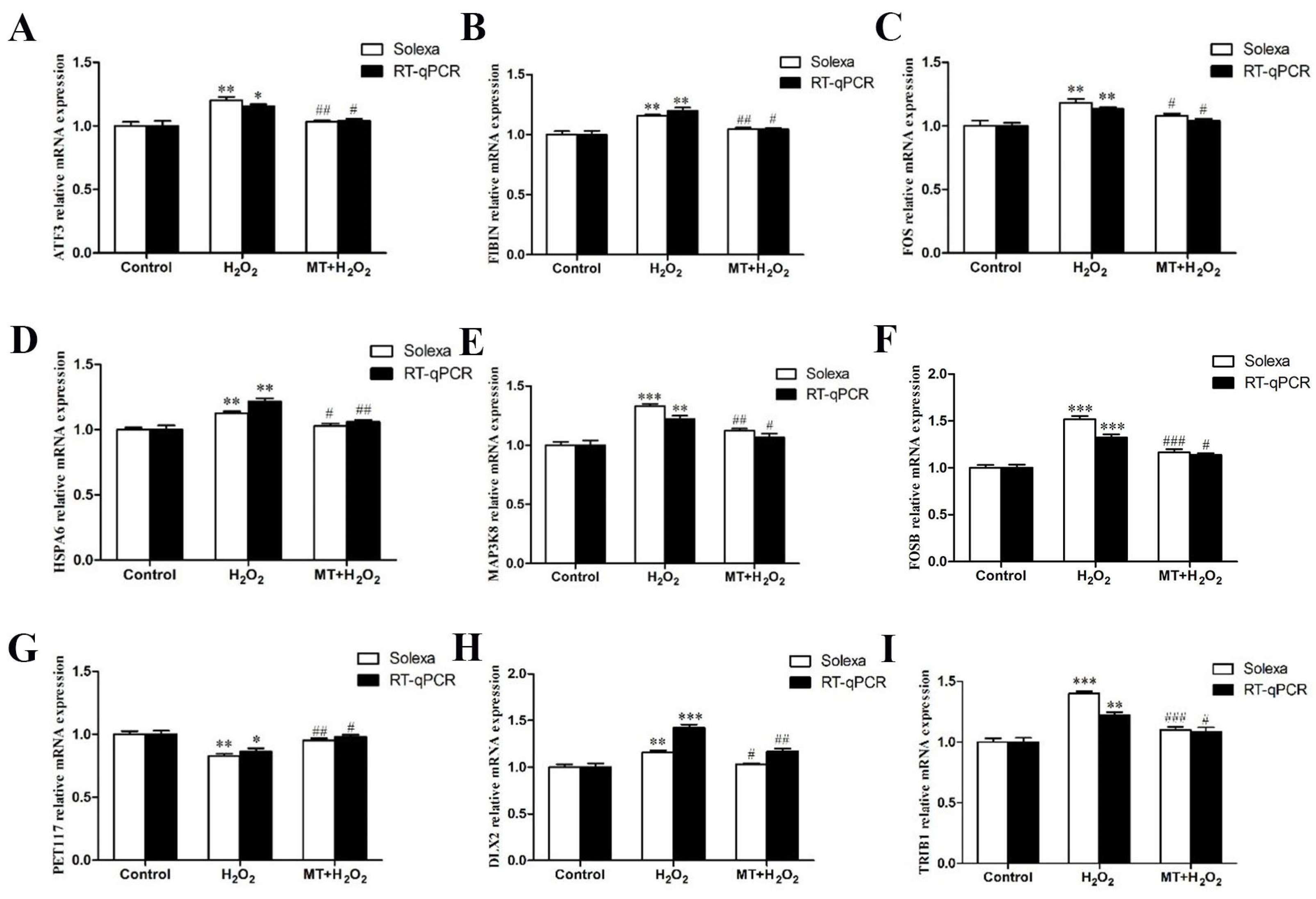
2.5. Real Time Quantitative Polymerase Chain Reaction (qRT-PCR) Analysis
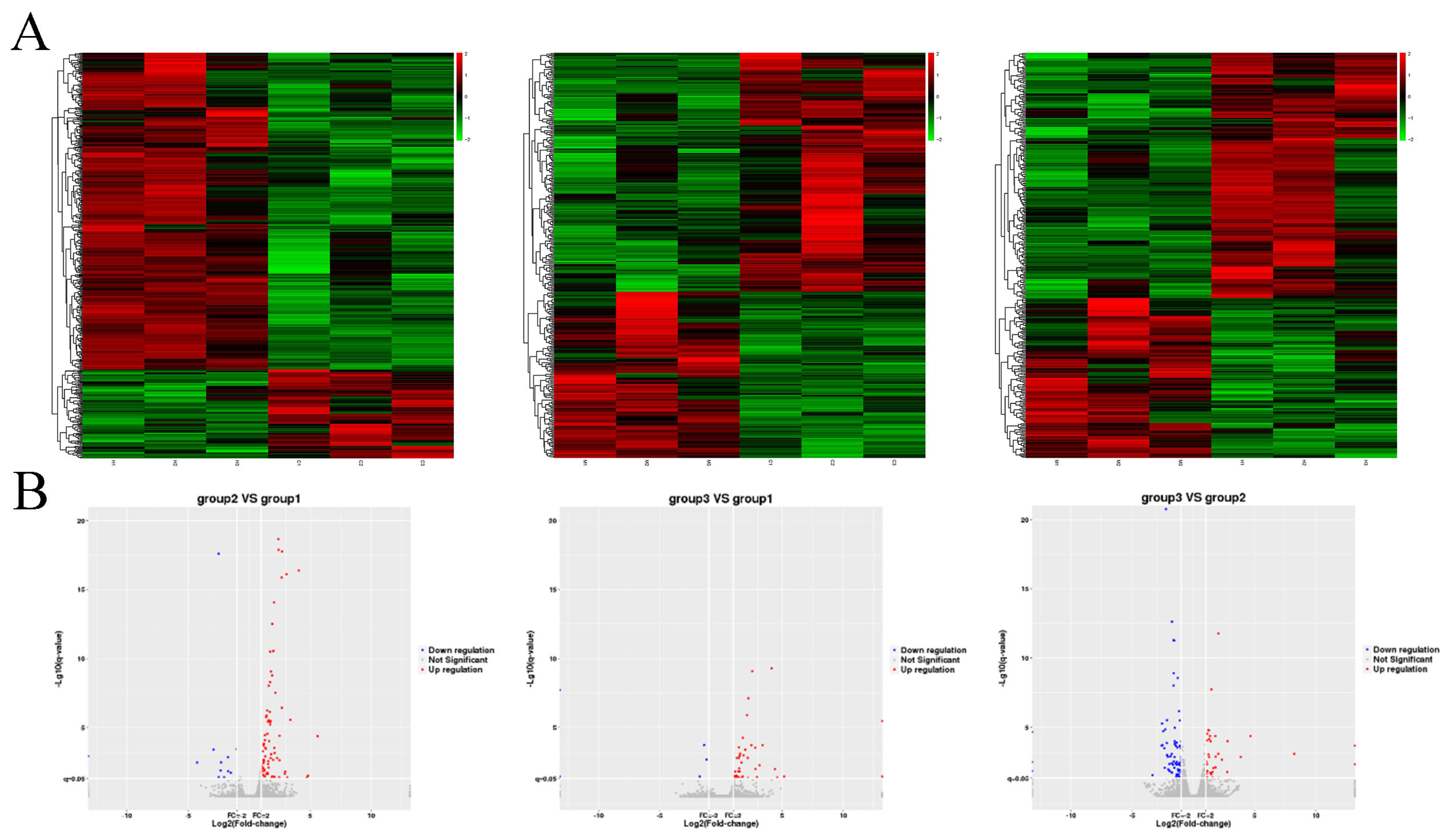
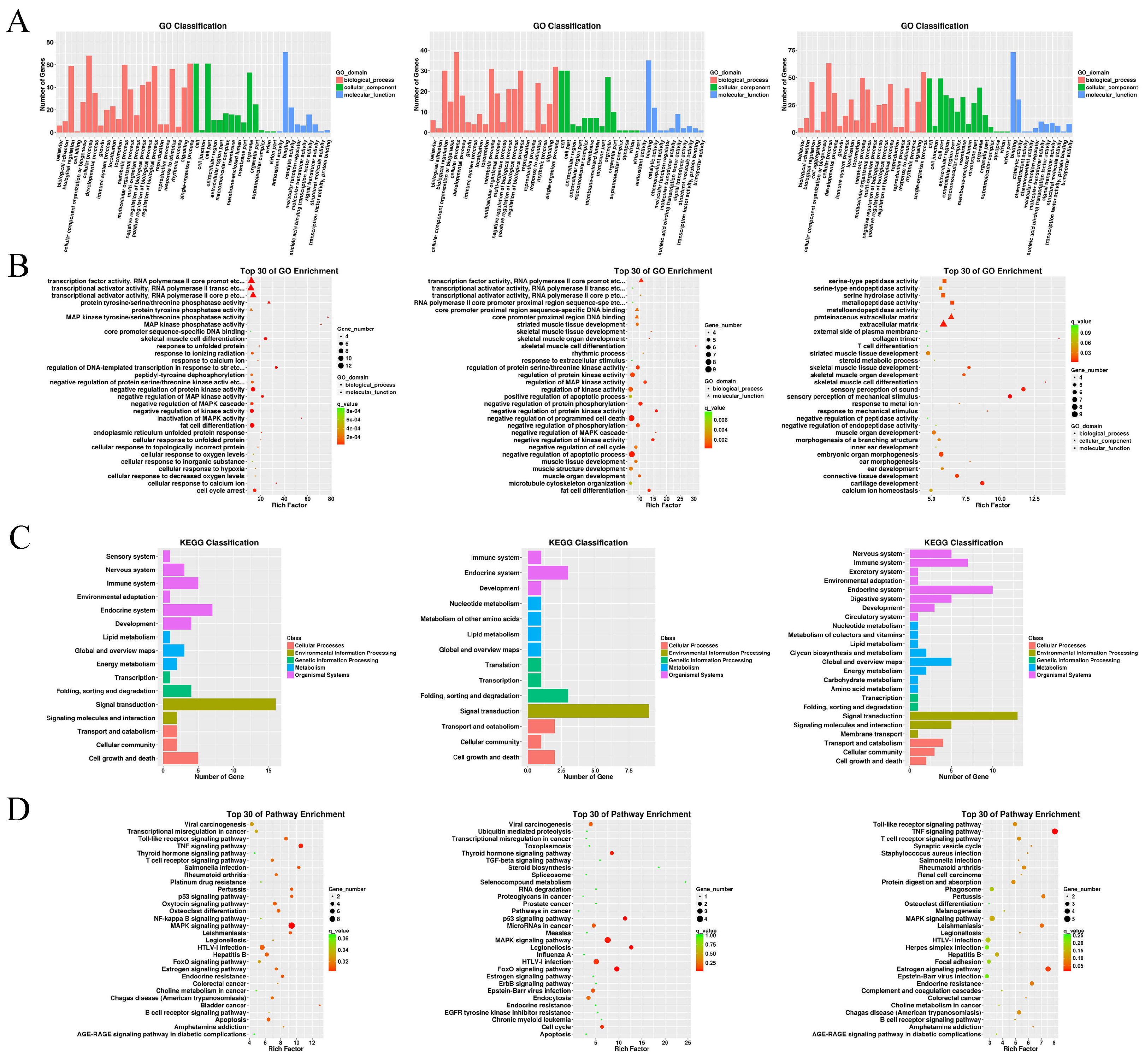
2.6. High-Throughput Sequencing Analysis
2.7. Western Blot Analysis
2.8. Overexpression Vector Construction
2.9. Statistical Analysis
3. Results
3.1. Melatonin Protects against H2O2-Induced Apoptosis in Sheep Granulosa Cells
3.2. High-Throughput Sequencing Analysis
3.3. Functional Enrichment and Signal Pathway Analysis
3.4. RT–qPCR Validation
3.5. Effect of Candidate Genes on Sheep Granulosa Cell Apoptosis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reiter, R.J.; Paredes, S.D.; Manchester, L.C.; Tan, D.X. Reducing oxidative/nitrosative stress: A newlydiscovered genre for melatonin. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 175–200. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tamura, H.; Tan, D.X.; Xu, X.Y. Melatonin and the circadian system: Contributions to successful female reproduction. Fertil. Steril. 2014, 102, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Tamura, H.; Takasaki, A.; Taketani, T.; Tanabe, M.; Kizuka, F.; Lee, L.; Tamura, I.; Maekawa, R.; Asada, H.; Yamagata, Y.; et al. Melatonin as a free radical scavenger in the ovarian follicle. Endocr. J. 2013, 60, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.R.; Bedaiwy, M.A.; Falcone, T. Analysis of apoptotic cell death, Bcl-2, and p53 protein expression in freshly fixed and cryopreserved ovarian tissue after exposure to warm ischemia. Fertil. Steril. 2006, 85, 1082–1092. [Google Scholar] [CrossRef]
- Barberino, R.S.; Menezes, V.G.; Ribeiro, A.E.; Palheta, R.C., Jr.; Jiang, X.; Smitz, J.E.; Matos, M.H.T. Melatonin protects against cisplatin-induced ovarian damage in mice via the MT1 receptor and antioxidant activity. Biol. Reprod. 2017, 96, 1244–1255. [Google Scholar] [CrossRef]
- Talpur, H.S.; Worku, T.; Rehman, Z.U.; Dad, R.; Bhattarai, D.; Bano, I.; Farmanullah; Liang, A.; He, C.; Yang, L. Knockdown of melatonin receptor 1 and induction of follicle-stimulating hormoneon the regulation of mouse granulosa cell function. Reprod. Biol. 2017, 17, 380–388. [Google Scholar] [CrossRef]
- Lin, T.; Lee, J.E.; Kang, J.W.; Oqani, R.K.; Cho, E.S.; Kim, S.B.; Jin, D., II. Melatonin supplementation during prolonged in vitro maturation improves the quality and development of poor-quality porcine oocytes via anti-oxidative andanti-apoptotic effects. Mol. Reprod. Dev. 2018, 85, 665–681. [Google Scholar] [CrossRef]
- He, Y.; Deng, H.; Jiang, Z.; Li, Q.; Shi, M.; Chen, H.; Han, Z. Effects of melatonin on follicular atresia and granulosa cell apoptosis in the porcine. Mol. Reprod. Dev. 2016, 83, 692–700. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, S. Melatonin promotes ubiquitination of phosphorylated pro-apoptotic protein Bcl2-interacting mediator of cell death-extra long (BimEL) in porcine granulosa cells. Int. J. Mol. Sci. 2018, 19, 3431–3446. [Google Scholar] [CrossRef]
- Fang, Y.; Deng, S.; Zhang, J.; Liu, H.; Li, Y.; Zhang, X.; Liu, Y. Melatonin-mediated development of ovine cumulus cells, perhaps by regulation of DNA methylation. Molecules 2018, 23, 494–506. [Google Scholar] [CrossRef]
- Riaz, H.; Yousuf, M.R.; Liang, A.; Hua, G.H.; Yang, L. Effect of melatonin on regulation of apoptosis and steroidogenesis in cultured buffalo granulosa cells. Anim. Sci. J. 2019, 90, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.-Q.; Zhang, Y.; Gao, Y.; Yang, X.-P.; Yang, Z.; Yang, Z.-J. The in vitro effects of melatonin and Cry gene on the secretion of estradiol from camel ovarian granulosa cells. Domest. Anim. Endocrinol. 2021, 74, 106497. [Google Scholar]
- Liu, Y.; Yang, Y.; Li, W.; Ao, H.; Zhang, Y.; Zhou, R.; Li, K. Effects of melatonin on the synthesis of estradiol and gene expression in pig granulosa cells. J. Pineal Res. 2019, 66, e12546. [Google Scholar] [CrossRef] [PubMed]
- Perego, M.C.; Bellitto, N.; Maylem, E.R.S.; Caloni, F.; Spicer, L.J. Effects of selected hormones and their combination on progesterone and estradiol production and proliferation of feline granulosa cells cultured in vitro. Theriogenology 2021, 168, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Liu, W.J.; Wu, C.J.; Ma, F.H.; Ahmad, S.; Liu, B.R.; Han, L.; Jiang, X.P.; Zhang, S.J.; Yang, L.G. Melatonin suppresses apoptosis and stimulates progesterone production by bovine granulosa cells via its receptors (MT1 and MT2). Theriogenology 2012, 78, 1517–1526. [Google Scholar] [CrossRef]
- Hao, E.-Y.; Wang, D.-H.; Chang, L.-Y.; Huang, C.-X.; Chen, H.; Yue, Q.-X.; Zhou, R.-Y.; Huang, R.-L. Melatonin regulates chicken granulosa cell proliferation and apoptosis by activating the mTOR signaling pathway via its receptors. Poult. Sci. 2020, 99, 6147–6162. [Google Scholar] [CrossRef]
- Wu, G.; Song, D.; Wei, Q.; Xing, J.; Shi, X.; Shi, F. Melatonin mitigates bisphenol A-induced estradiol production and proliferation by porcine ovarian granulosa cells in vitro. Anim. Reprod. Sci. 2018, 192, 91–98. [Google Scholar] [CrossRef]
- Nakamura, E.; Otsuka, F.; Terasaka, T.; Inagaki, K.; Hosoya, T.; Tsukamoto-Yamauchi, N.; Toma, K.; Makino, H. Melatonin counteracts BMP-6 regulation of steroidogenesis by rat granulosa cells. J. Steroid Biochem. Mol. Biol. 2014, 143, 233–239. [Google Scholar] [CrossRef]
- Fu, Y.; He, C.-J.; Ji, P.-Y.; Zhuo, Z.-Y.; Tian, X.-Z.; Wang, F.; Tan, D.-X.; Liu, G.-S. Effects of melatonin on the proliferation and apoptosis of sheep granulosa cells under thermal stress. Gene 2014, 15, 21090–21104. [Google Scholar] [CrossRef]
- Li, X.; Xu, G.; Li, Z.; Liu, H.; Ma, X.; Yang, L.; Zhang, P.; Zhao, J.; Wang, J.; Lu, W. Gonadotropin-Inhibiting Hormone Promotes Apoptosis of Bovine Ovary Granulosa Cells. Life Sci. 2021, 270, 119063. [Google Scholar] [CrossRef]
- Tian, X.; Wang, F.; He, C.; Zhang, L.; Tan, D.; Reiter, R.J.; Xu, J.; Ji, P.; Liu, G. Beneficial effects of melatonin on bovine oocytes maturation: A mechanistic approach. J. Pineal Res. 2014, 57, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.-M.; Tian, X.-Z.; Zhou, G.-B.; Wang, L.; Gao, C.; Zhu, S.-E.; Zeng, S.-M.; Tian, J.-H.; Liu, G.-S. Melatonin exists in porcine follicular fluid and improves in vitro maturation and parthenogenetic development of porcine oocytes. J. Pineal Res. 2010, 47, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Asma, A.; Marc-André, S. Melatonin Signaling Pathways Implicated in Metabolic Processes in Human Granulosa Cells (KGN). Int. J. Mol. Sci. 2022, 23, 2988. [Google Scholar] [CrossRef]
- Chen, Z.; Lei, L.; Wen, D.; Yang, L. Melatonin attenuates palmitic acid-induced mouse granulosa cells apoptosis via endoplasmic reticulum stress. J. Ovarian Res. 2019, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Li, S.; Zou, H.; Ji, D.; Lv, M.; Zhou, P.; Wei, Z.; Zhang, Z.; Cao, Y. Melatonin alleviates deoxynivalenol-induced apoptosis of human granulosa cells by reducing mutually accentuated FOXO1 and ER stress. Biol. Reprod. 2021, 105, 554–566. [Google Scholar] [CrossRef]
- Basini, G.; Simona, B.; Santini, S.E.; Grasselli, F. Reactive oxygen species and anti-oxidant defences in swine follicular fluids. Reprod. Fertil. Dev. 2008, 20, 269–274. [Google Scholar] [CrossRef]
- Agarwal, A.; Gupta, S.; Sharma, R.K. Role of oxidative stress in female reproduction. Reprod. Biol. Endocrinol. 2005, 3, 28–49. [Google Scholar] [CrossRef]
- Hao, E. Research on the Regulatory Mechanism of mTOR Signaling Pathway Mediated Melatonin to Delay Ovary Aginginlate-Phase Laying Hen. Ph.D. Thesis, HeBei Agricultural University, Baoding, China, 2020. [Google Scholar]
- Al-Shahat, A.; Hulail, M.A.E.; Soliman, N.M.M.; Khamis, T.; Fericean, L.M.; Arisha, A.H.; Moawad, R.S. Melatonin Mitigates Cisplatin-Induced Ovarian Dysfunction via Altering Steroidogenesis, Inflammation, Apoptosis, Oxidative Stress, and PTEN/PI3K/Akt/mTOR/AMPK Signaling Pathway in Female Rats. Pharmaceutics 2022, 14, 2769. [Google Scholar] [CrossRef]
- Shen, M.; Cao, Y.; Jiang, Y.; Wei, Y.; Liu, H. Melatonin protects mouse granulosa cells against oxidative damage by inhibiting FOXO1-mediated autophagy: Implication of an antioxidation-independent mechanism. Redox Biol. 2018, 18, 138–157. [Google Scholar] [CrossRef]
- Zheng, B.; Meng, J.; Zhu, Y.; Ding, M.; Zhang, Y.; Zhou, J. Melatonin enhances SIRT1 to ameliorate mitochondrial membrane damage by activating PDK1/Akt in granulosa cells of PCOS. J. Ovarian Res. 2021, 14, 152. [Google Scholar] [CrossRef]
- Yi, S.; Zheng, B.; Zhu, Y.; Cai, Y.; Sun, H.; Zhou, J. Melatonin ameliorates excessive PINK1/Parkin-mediated mitophagy by enhancing SIRT1 expression in granulosa cells of PCOS. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E91–E101. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.-L.; Zhang, X.; Zhou, J.-Q.; Li, C.-Y.; Yang, M.-H.; Liu, Z.-J.; Zhang, L.-L.; Deng, S.-L.; Zhang, L.; Shen, M.; et al. Melatonin Alleviates Hypoxia-Induced Apoptosis of Granulosa Cells by Reducing ROS and Activating MTNR1B-PKA-Caspase8/9 Pathway. Antioxidants 2021, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Garcia, C.; Sánchez, Á.; Contreras-Jurado, C.; Cales, C.; Barranquero, C.; Muñoz, M.; Merino, R.; Escudero, P.; Sanz, M.-J.; Osada, J.; et al. MAP3K8 Modulates Monocyte State and Atherogenesis in ApoE-/- Mice. Arter. Thromb. Vasc. Biol. 2017, 37, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Hebron, E.; Hope, C.; Kim, J.; Jensen, J.L.; Flanagan, C.; Bhatia, N.; Maroulakou, I.; Mitsiades, C.; Miyamoto, S.; Callander, N.; et al. MAP3K8 kinase regulates myeloma growth by cell-autonomous and non-autonomous mechanisms involving myeloma-associated monocytes/macrophages. Br. J. Haematol. 2013, 160, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Gianì, F.; Russo, G.; Pennisi, M.; Sciacca, L.; Frasca, F.; Pappalardo, F. Computational modeling reveals MAP3K8 as mediator of resistance to vemurafenib in thyroid cancer stem cells. Bioinformatics 2019, 35, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Wongkrasant, P.; Pongkorpsakol, P.; Chitwattananont, S.; Satianrapapong, W.; Tuangkijkul, N.; Muanprasat, C. 4 Fructo-oligosaccharides alleviate inflammation- associated apoptosis of GLP-1 secreting L cells via inhibition of iNOS and cleaved caspase-3 expression. J. Pharm. Sci. 2020, 143, 65–73. [Google Scholar] [CrossRef]
- Yao, L.; Yang, L.; Song, H.; Liu, T.; Yan, H. MicroRNA miR-29c-3p modulates FOS expression to repress EMT and cell proliferation while induces apoptosis in TGF-β2-treated lens epithelial cells regulated by lncRNA KCNQ1OT1. Biomed. Pharmacother. 2020, 129, 110290. [Google Scholar] [CrossRef]

| Genes | Primer Sequence (5’–3’) | Size (bp) |
|---|---|---|
| FOSB | F: TCCGCCGAGTCTCAGTATCTGTC R: TGGAGGTCCTGGCTGGTTGTG | 146 |
| TRIB1 | F: GACGGGAGCCTTCGGAGAGAC R: GTCACTGTCCTCCTGGTACTCTGG | 140 |
| DLX2 | F: TTGACAGTCTGGTGGCTGATATGC R: GCTTGTGGAGGCTGCTGCTG | 129 |
| PET117 | F: GAGACGCTCCAAGGTGGTGTTG R: GTCCTGCCGCTGCTTCAGATG | 85 |
| MAP3K8 | F: ACAACGAGGAGGAGCGGTCTG R: ACGCTTCGCAGTGTTGGATATGT | 131 |
| HSPA6 | F: AAGGAGACGGCGGAGGCTTAC R:GCTGCGAGTCGTTGAAGTAGGC | 82 |
| FOS | F: CACCGACCTGCCTGCAAGATC R: GCTCCATGCTGCCGACACTC | 193 |
| FIBIN | F:CAGGCTCAACGAGGACTTCTTGG R:CATGGCTCCTGATGGTGAGTGC | 125 |
| ATF3 | F: GAAGTCAGTGCCTCTGCCATCG R: GGCGAACCTCAGCTCTTCCTTG | 108 |
| Bcl2 | F: CGCATCGTGGCCTTCTTT R: CGGTTCAGGTACTCGGTCATC | 113 |
| Bax | F: CGAGTGGCGGCTGAAAT R: GGTCTGCCATGTGGGTGTC | 237 |
| β-actin | F: GCAAAGACCTCTACGCCAAC R: GGGCAGTGATCTCTTTCTGC | 90 |
| Gene | Full Genetic Name | p-Value | Error Discovery Rate | Log2 | Up/ Down |
|---|---|---|---|---|---|
| UNC13C | unc-13 homolog C | 4.03 × 10−16 | 1.69 × 10−12 | 2.035441 | UP |
| STAR | steroidogenic acute regulatory protein | 1.13 × 10−11 | 1.88 × 10−08 | 1.465299 | UP |
| MMP9 | matrix metallopeptidase 9 | 6.34 × 10−08 | 4.42 × 10−05 | 1.785441 | UP |
| PET117 | collagen type II alpha 1 chain | 6.12 × 10−08 | 4.42 × 10−05 | 4.656607 | UP |
| HSPA6 | heat shock protein family A (Hsp70) 6member 6 | 2.68 × 10−29 | 4.49 × 10−25 | −2.76246 | DOWN |
| FGL2 | fibrinogen like 2 | 2.12 × 10−25 | 1.77 × 10−21 | −2.2515 | DOWN |
| TRIB1 | tribbles pseudokinase 1 | 4.25 × 10−17 | 2.37 × 10−13 | −1.75357 | DOWN |
| FOS | Fos proto-oncogene, AP-1 transcription factor subunit | 1.94 × 10−15 | 5.43 × 10−12 | −1.57275 | DOWN |
| FOSB | Fos proto-oncogene B, AP-1 transcription factor subunit | 5.24 × 10−13 | 1.25 × 10−09 | −2.60732 | DOWN |
| MAK3P8 | mitogen-activated protein kinase kinase kinase 8 | 1.32 × 10−12 | 2.77 × 10−09 | −3.29209 | DOWN |
| SFRP4 | secreted frizzled related protein 4 | 5.21 × 10−12 | 9.70 × 10−09 | −1.6262 | DOWN |
| JUN | Jun proto-oncogene, AP-1 transcription factor subunit | 4.70 × 10−10 | 7.15 × 10−07 | −1.1781 | DOWN |
| ATF3 | Activating transcription factor 3 | 5.95 × 10−08 | 4.42 × 10−05 | −1.347907 | DOWN |
| FIBIN | fin bud initiation factor homolog (zebrafish) | 2.54 × 10−09 | 3.28 × 10−06 | −1.1054 | DOWN |
| STC1 | stanniocalcin 1 | 1.12 × 10−07 | 7.49 × 10−05 | −1.283589 | DOWN |
| TGM2 | transglutaminase 2 | 1.43 × 10−07 | 9.19 × 10−05 | −1.092309 | DOWN |
| DLX2 | distal-less homeobox 2 | 4.64 × 10−09 | 5.55 × 10−06 | −2.56006 | DOWN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, B.; Li, X.; Zhao, Z.; Cao, Y.; Liu, X.; Liu, Z.; Ma, H.; Lu, W. Melatonin Protects the Apoptosis of Sheep Granulosa Cells by Suppressing Oxidative Stress via MAP3K8 and FOS Pathway. Genes 2023, 14, 1067. https://doi.org/10.3390/genes14051067
Zhai B, Li X, Zhao Z, Cao Y, Liu X, Liu Z, Ma H, Lu W. Melatonin Protects the Apoptosis of Sheep Granulosa Cells by Suppressing Oxidative Stress via MAP3K8 and FOS Pathway. Genes. 2023; 14(5):1067. https://doi.org/10.3390/genes14051067
Chicago/Turabian StyleZhai, Bo, Xu Li, Zhongli Zhao, Yang Cao, Xinxin Liu, Zheng Liu, Huihai Ma, and Wenfa Lu. 2023. "Melatonin Protects the Apoptosis of Sheep Granulosa Cells by Suppressing Oxidative Stress via MAP3K8 and FOS Pathway" Genes 14, no. 5: 1067. https://doi.org/10.3390/genes14051067
APA StyleZhai, B., Li, X., Zhao, Z., Cao, Y., Liu, X., Liu, Z., Ma, H., & Lu, W. (2023). Melatonin Protects the Apoptosis of Sheep Granulosa Cells by Suppressing Oxidative Stress via MAP3K8 and FOS Pathway. Genes, 14(5), 1067. https://doi.org/10.3390/genes14051067






