The Lesson Learned from the Unique Evolutionary Story of Avirulence Gene AvrPii of Magnaporthe oryzae
Abstract
1. Introduction
2. Materials and Methods
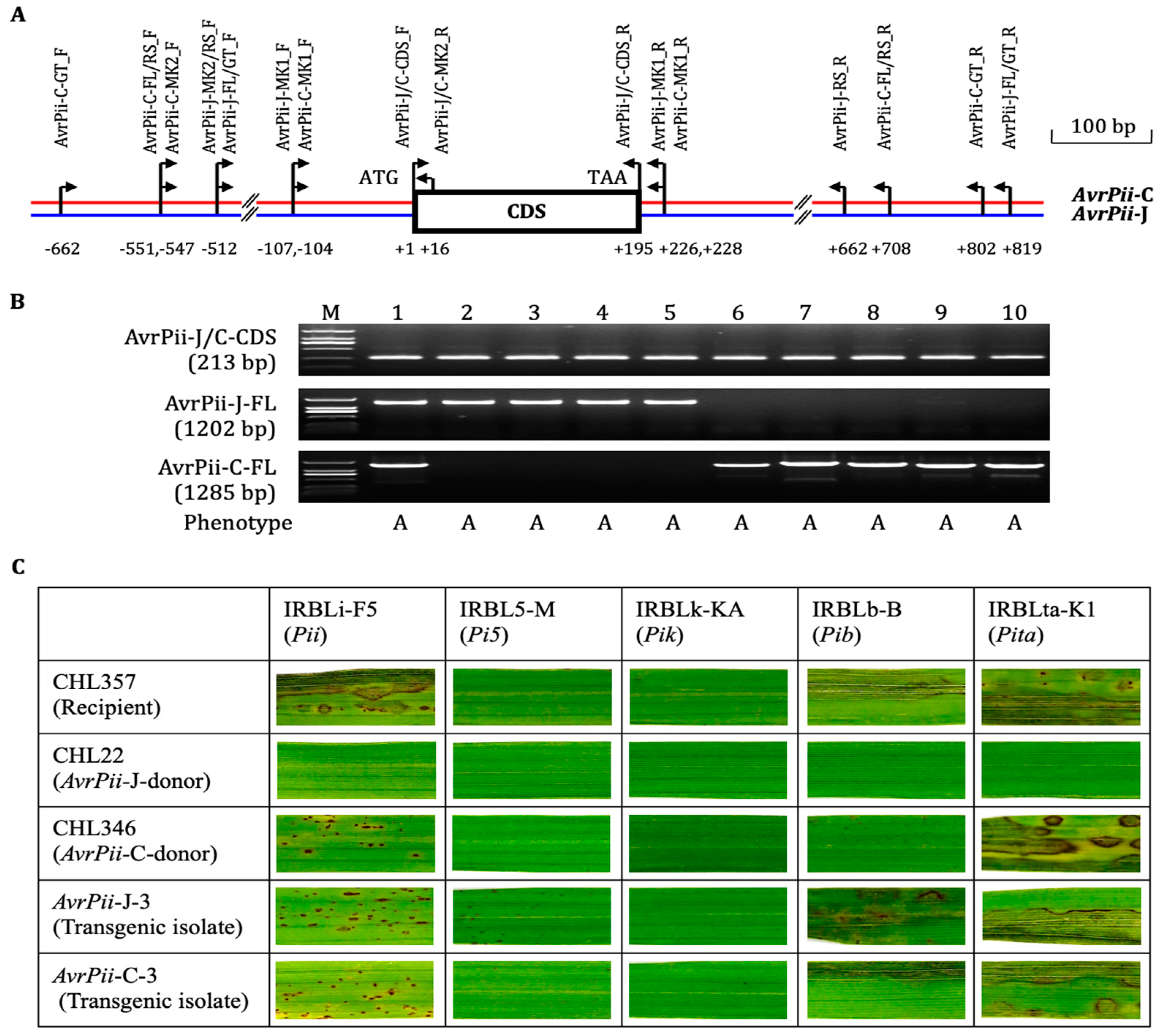
2.1. Discovery of a New Haplotype AvrPii-C
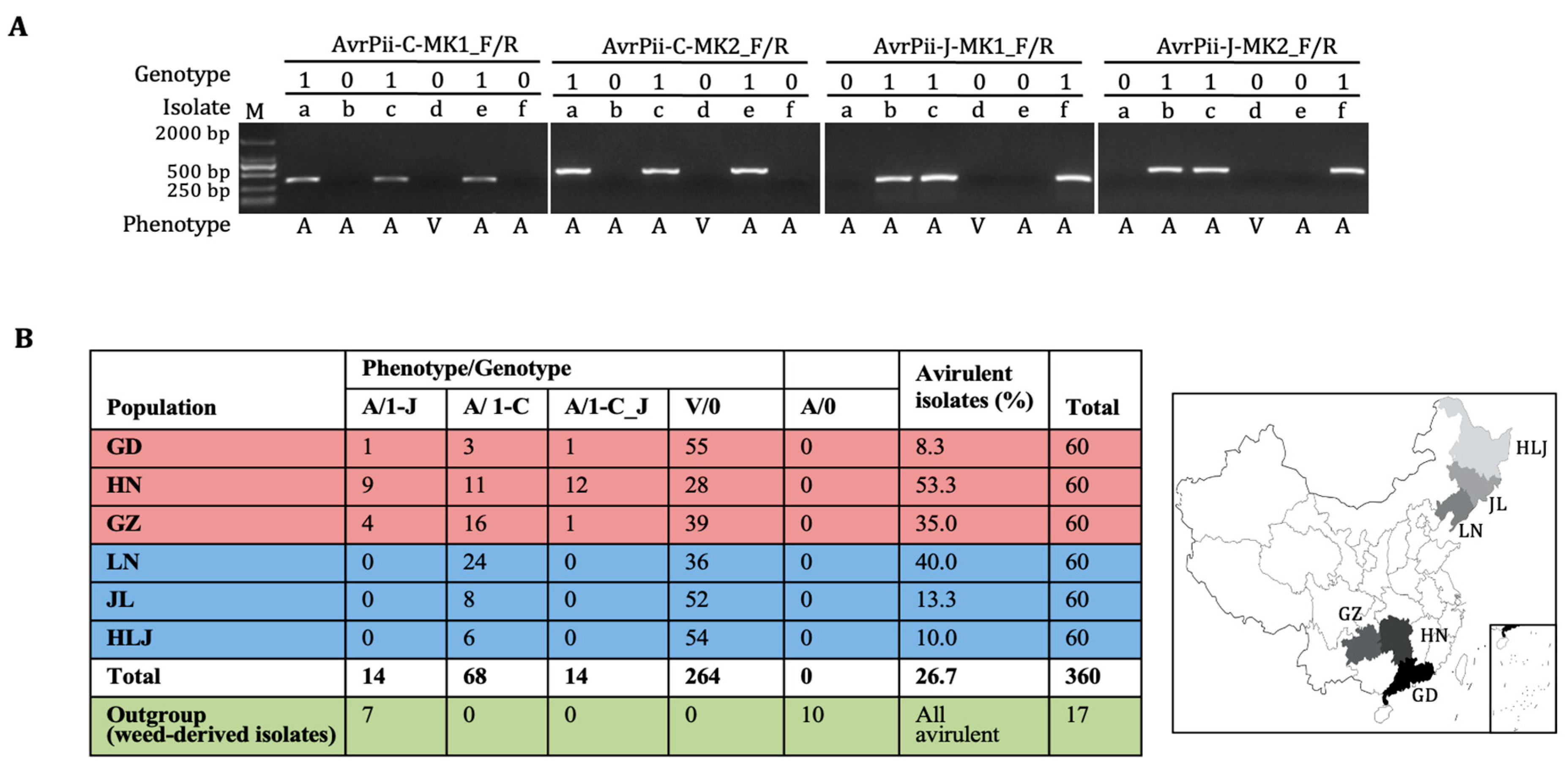
2.2. Functional Validation of Both Haplotypes
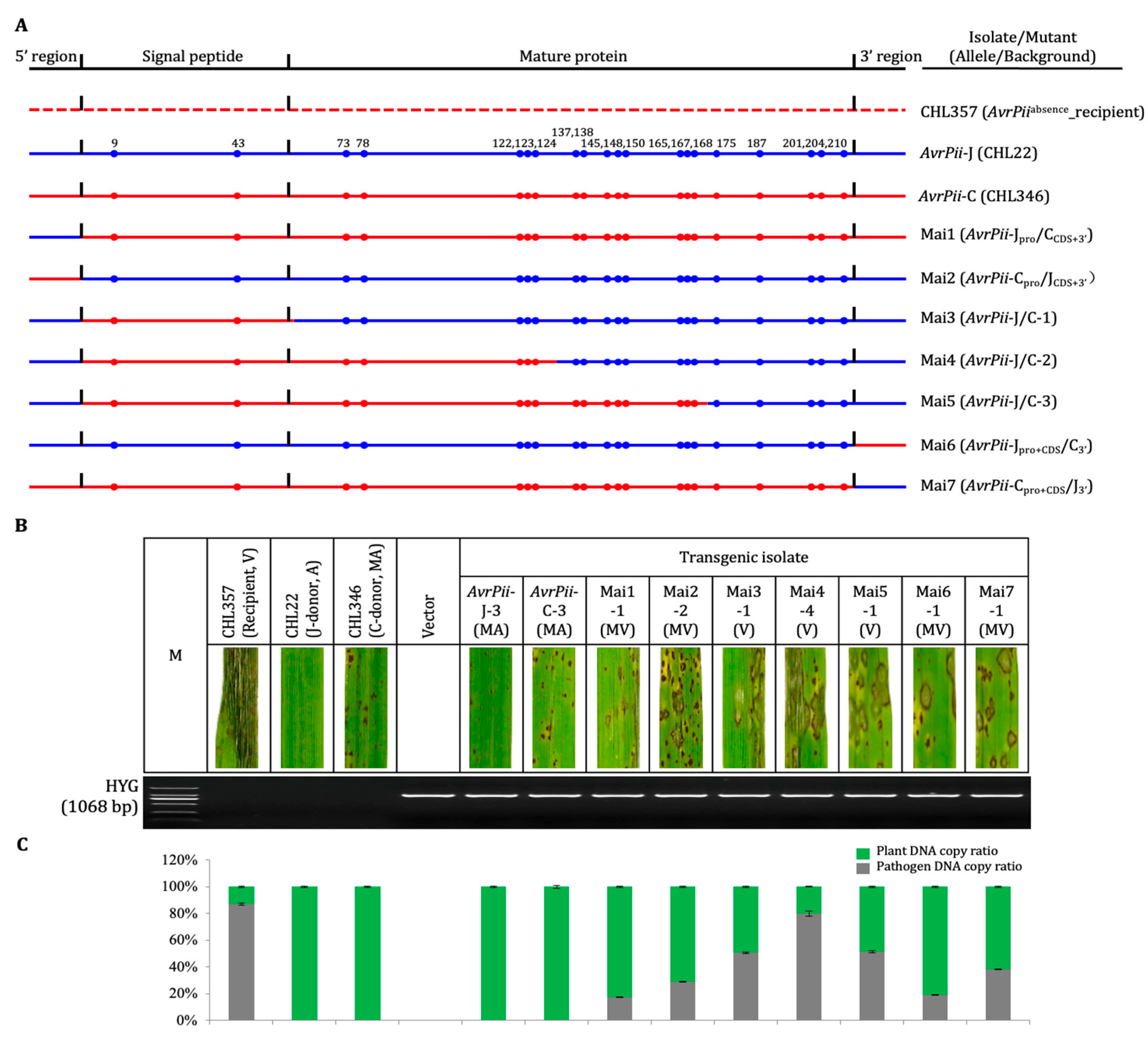
2.3. Construction and Characterization of Haplotype-Chimeric Mutant
2.4. Shape of Population Structure
2.5. Resequencing and Evolution Analysis
3. Results
3.1. Haplotypic Features of AvrPii
3.2. Performance of Haplotype-Chimeric Mutant
3.3. Population Structure of the AvrPii Family
3.4. Evolutionary and Regional Divergence of the AvrPii Family
4. Discussion
4.1. The Lesson Learned from Case Study on Population Structures of the AvrPii Family
4.2. The Lesson Learned from Evolutionary Story of the AvrPii Family
5. Conclusions
6. Patent
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, S. Retrospect and prospect of rice breeding in China. In Rice Breeding and Cultivar Genealogies in China during 1986 to 2005; Wan, J., Ed.; China Agr. Press: Beijing, China, 2010; pp. 1–23. [Google Scholar]
- Huang, Z.; Wang, J.; Zhang, Y.; Yao, Y.; Huang, L.; Yang, X.; Wang, L.; Pan, Q. Dynamics of race structures of Pyricularia oryzae populations across 18 seasons in Guangdong province, China. Plant Dis. 2021, 105, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wang, Q.; Jia, Y.; Bi, Y.; Li, C.; Fan, H.; Li, J. Selection and mutation of the avirulence gene AvrPii of the rice blast fungus Magnaporthe oryzae. Plant Pathol. 2019, 68, 127–134. [Google Scholar] [CrossRef]
- Peng, Z.; Li, L.; Wu, S.; Chen, X.; Shi, Y.; He, Q.; Shu, F.; Zhang, W.; Sun, P.; Deng, H.; et al. Frequencies and variation of Magnaporthe oryzae avirulence genes in Hunan province, China. Plant Dis. 2021, 105, 3829–3834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Yao, Y.; Jin, X.; Correll, J.; Wang, L.; Pan, Q. Dynamics of race structures of the rice blast pathogen populations in Heilongjiang province, China from 2006 through 2015. Plant Dis. 2019, 103, 2759–2763. [Google Scholar] [CrossRef]
- Chai, R.; Wang, J.; Wang, X.; Wen, J.; Liang, Z.; Ye, X.; Zhang, Y.; Yao, Y.; Zhang, J.; Zhang, Y.; et al. The Pid family has been diverged into Xian and Geng type resistance genes against rice blast disease. Genes 2022, 13, 891. [Google Scholar] [CrossRef]
- Wu, W.; Wang, L.; Zhang, S.; Li, Z.; Zhang, Y.; Lin, F.; Pan, Q. Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant-Microbe Interac. 2014, 27, 759–769. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Wu, W.; He, L.; Yang, X.; Pan, Q. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 2015, 5, 11642. [Google Scholar] [CrossRef]
- Chen, C.; Chen, M.; Hu, J.; Zhang, W.; Zhong, Z.; Jia, Y.; Allaux, L.; Fournier, E.; Tharreau, D.; Wang, G.; et al. Sequence variation and recognition specificity of the avriulence gene AvrPiz-t in Magnaporthe oryzae field populations. Fungal Genom. Biol. 2014, 4, 1. [Google Scholar]
- Flor, H. Current status of the gene for gene concept. Ann. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Sirisathaworn, T.; Srirat, T.; Longya, A.; Jantasuriyarat, C. Evaluation of mating type distribution and genetic diversity of three Magnaporthe oryzae avirulence genes, PWL2, AvrPii and AvrPiz-t, in Thailand rice blast isolates. Agric. Nat. Resour. 2017, 51, 7–14. [Google Scholar]
- Zhang, Y.; Zhu, Q.; Yao, Y.; Zhao, Z.; Correll, J.; Wang, L.; Pan, Q. The race structure of the rice blast pathogen across southern and northeastern China. Rice 2017, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Siatoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Jia, Y.; Peng, Z.; Shi, Y.; He, Q.; Shu, F.; Zhang, W.; Zhang, Z.; Deng, H. Characterization of molecular identity and pathogenicity of rice blast fungus in Hunan province of China. Plant Dis. 2017, 101, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jia, Y.; Wang, Y.; Sun, G. A rapid survey of avirulence genes in field isolates of Magnaporthe oryzae. Plant Dis. 2020, 104, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, H.; Yanoria, M.; Ebron, L.; Hayashi, N.; Ando, I.; Kata, H.; Imbe, T.; Khush, G. Development of monogenic lines of rice for rice blast resistance. Breed. Sci. 2000, 50, 229–234. [Google Scholar] [CrossRef]
- Pan, Q.; Wang, L.; Ikehashi, H.; Tanisaka, T. Identification of a new blast resistance gene in the Xian rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 1996, 86, 1071–1075. [Google Scholar] [CrossRef]
- Zhai, C.; Zhang, Y.; Yao, N.; Lin, F.; Liu, Z.; Dong, Z.; Wang, L.; Pan, Q. Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS ONE 2014, 9, e98067. [Google Scholar] [CrossRef]
- Lynch, M.; Crease, T. The analysis of population survey data on DNA sequence variation. Mol. Biol. Evol. 1990, 7, 377–394. [Google Scholar]
- Nei, M. Molecular Evolutionary Genetics; Columbia Univ. Press: New York, NY, USA, 1987. [Google Scholar]
- Fu, Y.; Li, W. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar]
- Han, G. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L. Genetics and the understanding of selection. Nat. Rev. Genet. 2009, 10, 83–93. [Google Scholar] [CrossRef] [PubMed]
- de Vries, S.; Stukenbrock, E.; Rose, L. Rapid evolution in plant-microbe interactions-and evolutionary genomics perspective. New Phytol. 2020, 226, 1256–1262. [Google Scholar] [CrossRef]
- Fujisaki, K.; Abe, Y.; Ito, A.; Saitoh, H.; Yoshida, K.; Kanzaki, H.; Kanzaki, E.; Utsushi, H.; Yamashita, T.; Kamoun, S.; et al. Rice Exo70 interacts with a fungal effector, AvrPii, and is required for AvrPii-triggered immunity. Plant J. 2015, 83, 875–887. [Google Scholar] [CrossRef]
- Kawasaki-Tanaka, A.; Hayashi, N.; Yanagihara, S.; Fukuta, Y. Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Japan. Plant Dis. 2016, 100, 816–823. [Google Scholar] [CrossRef]
- Khan, M.; Ali, M.; Monsur, M.; Kawasaki-Tanaka, A.; Hayashi, N.; Yanagihara, S.; Obara, M.; Mia, M.; Latif, M.; Fukuta, Y. Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Bangladesh. Plant Dis. 2016, 100, 2025–2033. [Google Scholar] [CrossRef]
- Fukuta, Y.; Telebanco-Yanoria, M.; Hayashi, N.; Yanagihara, S.; Machungo, C.; Makihara, D. Pathogenicities of rice blast (Pyricularia oryzae Cavara) isolates from Kenya. Plant Dis. 2019, 103, 3188. [Google Scholar] [CrossRef] [PubMed]
- Nguyet, N.; Long, H.; Ngoc, N.; Nhai, N.; Thuy, N.; Hayashi, N.; Fukuta, Y. Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Vietnam. Plant Dis. 2020, 104, 381–387. [Google Scholar] [CrossRef]
- Chuma, I.; Isobe, C.; Hotta, Y.; Ibaragi, K.; Futamata, N.; Kusaba, M.; Yoshida, K.; Terauchi, R.; Fujita, Y.; Nakayashiki, H.; et al. Multiple translocations of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 2011, 7, e1002147. [Google Scholar] [CrossRef]
- Dai, Y.; Jia, Y.; Correll, J.; Wang, X.; Wang, Y. Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe oryzae. Fungal Genet. Biol. 2010, 47, 973–980. [Google Scholar] [CrossRef]
- Khang, C.; Park, S.; Lee, Y.; Valent, B.; Kang, S. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant-Microbe Interac. 2008, 21, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Q.; Li, C.; Bi, Y.; Fu, X.; Wang, R. Novel haplotypes and networks of AvrPik alleles in Magnaporthe oryzae. BMC Plant Biol. 2019, 19, 204. [Google Scholar] [CrossRef] [PubMed]
- Longya, A.; Chaipanya, C.; Franceschetti, M.; Maidment, J.; Banfiled, M.; Jantasuriyarat, C. Gene duplication and mutation in the emergence of a novel aggressive allele of the AvrPik effector in the rice blast fungus. Mol. Plant-Microbe Interac. 2019, 32, 740–749. [Google Scholar] [CrossRef]
- Praz, C.; Bourras, S.; Zeng, F.; Sanchez-Martin, J.; Menardo, F.; Xue, M.; Yang, L.; Roffler, S.; Boni, R.; Herren, G.; et al. AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytol. 2017, 213, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Ashizawa, T.; Hirayae, K.; Moriwaki, J.; Sone, T.; Sonoda, R.; Noguchi, M.; Nagashima, S.; Ishikawa, K.; Arai, M. One of two paralogs of AvrPita1 is functional in Japanese rice blast isolates. Phytopathology 2010, 100, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, H.; Duan, G.; Huang, Y.; Liu, S.; Fang, Z.; Wu, E.; Shang, L.; Zhan, J. The Phytophthora infestans Avr2 effector escapes R2 recognition through effector discording. Mol. Plant-Microbe Interac. 2020, 33, 921–931. [Google Scholar] [CrossRef]
- Dodds, P.; Thrall, P. Recognition events and host-pathogen co-evolution in gene-for-gene resistance to flax rust. Funct. Plant Biol. 2009, 36, 395–408. [Google Scholar] [CrossRef]
- Schürch, S.; Linde, C.; Knogge, W.; Jackson, L.; McDonald, B. Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol. Plant-Microbe Interac. 2004, 17, 1114–1125. [Google Scholar] [CrossRef]
- Daugherty, M.; Malik, H. Rules of engagement: Molecular insights from host-virus arms races. Annu. Rev. Genet. 2012, 46, 677–700. [Google Scholar] [CrossRef]
- Lin, L. Identification of Rice Blast Resistance Genes in Current Cultivars in Liaoning and Heilongjiang Provinces, China. Master’s Dissertation, South China Agricultural University, Guangzhou, China, June 2021. (In Chinese). [Google Scholar]
- Ye, X. Identifying the Composition of Rice Blast Resistance Genes and Mining New Allelic Genes of Backbone Rice Cultivars in Guangdong Province and Guangxi Zhuang Autonomous Region. Master’s Dissertation, South China Agricultural University, Guangzhou, China, June 2021. (In Chinese). [Google Scholar]
- Zeng, S. Identification of Rice Blast Resistance Genes in the Key Parental Cultivars. Master’s Dissertation, South China Agricultural University, Guangzhou, China, June 2020. (In Chinese). [Google Scholar]
- China National Rice Research Institute. Regionalization of Rice Cropping in China; Zhejiang Sci. Tech. Press: Hangzhou, China, 1988. (In Chinese) [Google Scholar]
| Comparable Group a | S b | π c | D* d | F* | Ka e | Ks f | Ka/Ks |
|---|---|---|---|---|---|---|---|
| All AvrPii (∑110) | |||||||
| Entire coding region | 20 | 0.037 | 1.74 β | 2.65 β | 193.8 | 357.6 | 0.54 |
| Non-signal peptide region | 18 | 0.045 | 1.68 α | 2.59 β | 246.8 | 434.8 | 0.57 |
| Signal peptide region | 2 | 0.013 | 0.68 | 1.11 | 54.9 | 166.0 | 0.33 |
| Haplotype AvrPii-J (∑28) | |||||||
| Entire coding region | 0 | 0.000 | 0.00 | 0.00 | 0.0 | 0.0 | Ka = Ks |
| Non-signal peptide region | 0 | 0.000 | 0.00 | 0.00 | 0.0 | 0.0 | Ka = Ks |
| Signal peptide region | 0 | 0.000 | 0.00 | 0.00 | 0.0 | 0.0 | Ka = Ks |
| Haplotype AvrPii-C (∑82) | |||||||
| Entire coding region | 1 | 0.002 | 0.51 | 0.79 | 8.1 | 0.0 | Ka > Ks |
| Non-signal peptide region | 1 | 0.002 | 0.51 | 0.79 | 11.0 | 0.0 | Ka > Ks |
| Signal peptide region | 0 | 0.000 | 0.00 | 0.00 | 0.0 | 0.0 | Ka = Ks |
| Regional group (∑110) | |||||||
| South (AvrPii-J, -C; ∑72) | 20 | 0.045 | 1.72 β | 3.04 β | 100.8 | 192.2 | 0.52 |
| North (AvrPii-C; ∑38) | 0 | 0.000 | 0.00 | 0.00 | 0.0 | 0.0 | Ka = Ks |
| Outgroup (∑7) | |||||||
| Weed-derived isolates (AvrPii-J) | 0 | 0.000 | 0.00 | 0.00 | 0.0 | 0.0 | Ka = Ks |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wu, W.; Zhang, Y.; Li, C.; Wang, J.; Wen, J.; Zhang, S.; Yao, Y.; Lu, W.; Zhao, Z.; et al. The Lesson Learned from the Unique Evolutionary Story of Avirulence Gene AvrPii of Magnaporthe oryzae. Genes 2023, 14, 1065. https://doi.org/10.3390/genes14051065
Wang X, Wu W, Zhang Y, Li C, Wang J, Wen J, Zhang S, Yao Y, Lu W, Zhao Z, et al. The Lesson Learned from the Unique Evolutionary Story of Avirulence Gene AvrPii of Magnaporthe oryzae. Genes. 2023; 14(5):1065. https://doi.org/10.3390/genes14051065
Chicago/Turabian StyleWang, Xing, Weihuai Wu, Yaling Zhang, Cheng Li, Jinyan Wang, Jianqiang Wen, Shulin Zhang, Yongxiang Yao, Weisheng Lu, Zhenghong Zhao, and et al. 2023. "The Lesson Learned from the Unique Evolutionary Story of Avirulence Gene AvrPii of Magnaporthe oryzae" Genes 14, no. 5: 1065. https://doi.org/10.3390/genes14051065
APA StyleWang, X., Wu, W., Zhang, Y., Li, C., Wang, J., Wen, J., Zhang, S., Yao, Y., Lu, W., Zhao, Z., Zhan, J., & Pan, Q. (2023). The Lesson Learned from the Unique Evolutionary Story of Avirulence Gene AvrPii of Magnaporthe oryzae. Genes, 14(5), 1065. https://doi.org/10.3390/genes14051065








