PTRH2 Gene Variants: Recent Review of the Phenotypic Features and Their Bioinformatics Analysis
Abstract
1. Introduction
2. Methodology
Structural Analysis of the Mutational Effects
3. Results
3.1. Case Presentation
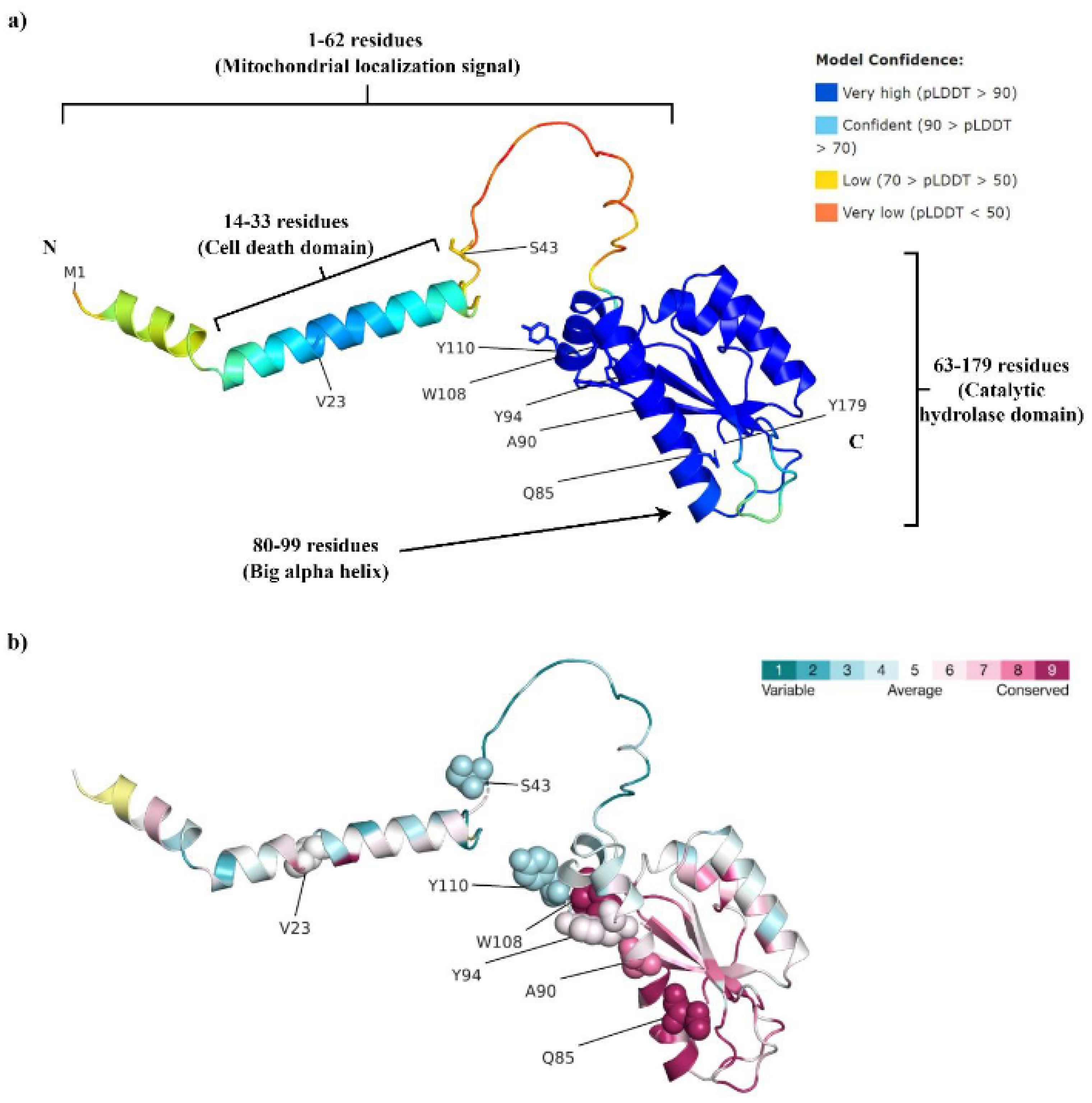
3.2. Structural Analysis of the Mutational Effects
3.2.1. V23A
3.2.2. S43Kfs*11
3.2.3. Q85P
3.2.4. A90Gfs*13
3.2.5. Y94N
3.2.6. W108*
3.2.7. E110*
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, H.; Matter, M.L.; Issa-Jahns, L.; Jijiwa, M.; Kraemer, N.; Musante, L.; de la Vega, M.; Ninnemann, O.; Schindler, D.; Damatova, N.; et al. Mutations in PTRH2 cause novel infantile-onset multisystem disease with intellectual disability, microcephaly, progressive ataxia, and muscle weakness. Ann. Clin. Transl. Neurol. 2014, 1, 1024–1035. [Google Scholar] [CrossRef]
- Picker-Minh, S.; Mignot, C.; Doummar, D.; Hashem, M.; Faqeih, E.; Josset, P.; Dubern, B.; Alkuraya, F.S.; Kraemer, N.; Kaindl, A.M. Phenotype variability of infantile-onset multisystem neurologic, endocrine, and pancreatic disease IMNEPD. Orphanet J. Rare Dis. 2016, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Sharkia, R.; Shalev, S.A.; Zalan, A.; Marom-David, M.; Watemberg, N.; Urquhart, J.E.; Daly, S.B.; Bhaskar, S.S.; Williams, S.G.; Newman, W.G.; et al. Homozygous mutation in PTRH2 gene causes progressive sensorineural deafness and peripheral neuropathy. Am. J. Med. Genet. A 2017, 173, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Le, C.; Prasad, A.N.; Rupar, C.A.; Debicki, D.; Andrade, A.; Prasad, C. Infantile-onset multisystem neurologic, endocrine, and pancreatic disease: Case and review. Can. J. Neurol. Sci. 2019, 46, 459–463. [Google Scholar] [CrossRef]
- Parida, P.; Dubbudu, A.; Biswal, S.R.; Sharawat, I.K.; Panda, P.K. Diabetes mellitus in an adolescent girl with intellectual disability caused by novel single base pair duplication in the PTRH2 gene: Expanding the clinical spectrum of IMNEPD. Brain Dev. 2021, 43, 314–319. [Google Scholar] [CrossRef]
- Charles Bronson, S.; Suresh, E.; Kumar, S.S.A.S.; Mythili, C.; Shanmugam, A. A novel synergistic association of variants in PTRH2 and KIF1A relates to a syndrome of hereditary axonopathy, outer hair cell dysfunction, intellectual disability, pancreatic lipomatosis, diabetes, cerebellar atrophy, and vertebral artery hypoplasia. Cureus 2021, 13, e13174. [Google Scholar] [CrossRef]
- Sharma, S.; Kaushik, S.; Sinha, M.; Kushwaha, G.S.; Singh, A.; Sikarwar, J.; Chaudhary, A.; Gupta, A.; Kaur, P.; Singh, T.P. Structural and functional insights into peptidyl-TRNA hydrolase. Biochim. Biophys. Acta 2014, 1844, 1279–1288. [Google Scholar] [CrossRef]
- Yao, X.; Jennings, S.; Ireland, S.K.; Pham, T.; Temple, B.; Davis, M.; Chen, R.; Davenport, I.; Biliran, H. The anoikis effector BIT1 displays tumor suppressive function in lung cancer cells. PLoS ONE 2014, 9, e101564. [Google Scholar] [CrossRef] [PubMed]
- Corpuz, J.M.; Waas, W.F.; Jan, Y.; Ruoslahti, E.; Schimmel, P.; Pascual, J. Crystal structure of a human peptidyl-TRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity. J. Biol. Chem. 2004, 279, 8111–8115. [Google Scholar] [CrossRef]
- Jan, Y.; Matter, M.; Pai, J.; Chen, Y.-L.; Pilch, J.; Komatsu, M.; Ong, E.; Fukuda, M.; Ruoslahti, E. A mitochondrial protein, BIT1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell 2004, 116, 751–762. [Google Scholar] [CrossRef]
- Griffiths, G.S.; Grundl, M.; Leychenko, A.; Reiter, S.; Young-Robbins, S.S.; Sulzmaier, F.J.; Caliva, M.J.; Ramos, J.W.; Matter, M.L. BIT-1 mediates integrin-dependent cell survival through activation of the NFkappaB pathway. J. Biol. Chem. 2011, 286, 14713–14723. [Google Scholar] [CrossRef]
- Yi, P.; Nguyên, D.T.; Higa-Nishiyama, A.; Auguste, P.; Bouchecareilh, M.; Dominguez, M.; Bielmann, R.; Palcy, S.; Liu, J.F.; Chevet, E. MAPK scaffolding by BIT1 in the Golgi complex modulates stress resistance. J. Cell Sci. 2010, 123 Pt 7, 1060–1072. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Regev, A.; Teichmann, S.A.; Lander, E.S.; Amit, I.; Benoist, C.; Birney, E.; Bodenmiller, B.; Campbell, P.; Carninci, P.; Clatworthy, M.; et al. Human Cell Atlas Meeting Participants. The human cell atlas. Elife 2017, 6, e27041. [Google Scholar] [CrossRef] [PubMed]
- Corpuz, A.D.; Ramos, J.W.; Matter, M.L. PTRH2: An adhesion regulated molecular switch at the nexus of life, death, and differentiation. Cell Death Discov. 2020, 6, 124. [Google Scholar] [CrossRef]
- Griffiths, G.S.; Doe, J.; Jijiwa, M.; Van Ry, P.; Cruz, V.; de la Vega, M.; Ramos, J.W.; Burkin, D.J.; Matter, M.L. BIT-1 is an essential regulator of myogenic differentiation. J. Cell Sci. 2015, 128, 1707–1717. [Google Scholar] [CrossRef]
- Doe, J.; Kaindl, A.M.; Jijiwa, M.; de la Vega, M.; Hu, H.; Griffiths, G.S.; Fontelonga, T.M.; Barraza, P.; Cruz, V.; Van Ry, P.; et al. PTRH2 gene mutation causes progressive congenital skeletal muscle pathology. Hum. Mol. Genet. 2017, 26, 1458–1464. [Google Scholar] [CrossRef] [PubMed]
- Duband, J.L.; Monier, F.; Delannet, M.; Newgreen, D. Epithelium-mesenchyme transition during neural crest development. Acta Anat. 1995, 154, 63–78. [Google Scholar] [CrossRef]
- Kerosuo, L.; Bronner-Fraser, M. What is bad in cancer is good in the embryo: Importance of EMT in neural crest development. Semin. Cell Dev. Biol. 2012, 23, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Ando, M.; Higuchi, Y.; Takeuchi, M.; Hashiguchi, A.; Takashima, H. The first case of infantile-onset multisystem neurologic, endocrine, and pancreatic disease caused by novel PTRH2 mutation in Japan. Neurol. Sci. 2022, 43, 2133–2136. [Google Scholar] [CrossRef]
- Sharkia, R.; Zalan, A.; Zahalka, H.; Kessel, A.; Asaly, A.; Al-Shareef, W.; Mahajnah, M. CLN8 gene compound heterozygous variants: A new case and protein bioinformatics analyses. Genes 2022, 13, 1393. [Google Scholar] [CrossRef]
- Suleman, M.; Yousafi, Q.; Ali, J.; Ali, S.S.; Hussain, Z.; Ali, S.; Waseem, M.; Iqbal, A.; Ahmad, S.; Khan, A.; et al. Bioinformatics analysis of the differences in the binding profile of the wild-type and mutants of the SARS-CoV-2 spike protein variants with the ACE2 receptor. Comput. Biol. Med. 2021, 138, 104936. [Google Scholar] [CrossRef] [PubMed]
- Alazami, A.M.; Patel, N.; Shamseldin, H.E.; Anazi, S.; Al-Dosari, M.S.; Alzahrani, F.; Hijazi, H.; Alshammari, M.; Aldahmesh, M.A.; Salih, M.A.; et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015, 10, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Abu Rayyan, A.; Kamal, L.; Casadei, S.; Brownstein, Z.; Zahdeh, F.; Shahin, H.; Canavati, C.; Dweik, D.; Jaraysa, T.; Rabie, G.; et al. genomic analysis of inherited hearing loss in the Palestinian population. Proc. Natl. Acad. Sci. USA 2020, 117, 20070–20076. [Google Scholar] [CrossRef]
- Khamirani, H.J.; Zoghi, S.; Dianatpour, M.; Jankhah, A.; Tabei, S.S.; Mohammadi, S.; Dastgheib, S.A. A novel PTRH2 missense mutation causing IMNEPD: A case report. Hum. Genome Var. 2021, 8, 23. [Google Scholar] [CrossRef]
- Becker, M.; Seneca, S.; Schierloh, U.; Witsch, M.; de Beaufort, C.; Scalais, E. Diabetes in a child with infantile onset multisystem neurological, endocrine and pancreatic disease (IMNEPD). Horm. Res. Paediatr. 2021, 94 (Suppl. 1), 228. Available online: https://researchportal.vub.be/en/publications/diabetes-in-a-child-with-infantile-onset-multisystem-neurological (accessed on 10 February 2023).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Porollo, A.; Meller, J. Prediction-based fingerprints of protein-protein interactions. Proteins 2007, 66, 630–645. [Google Scholar] [CrossRef]
- Tubiana, J.; Schneidman-Duhovny, D.; Wolfson, H.J. ScanNet: An interpretable geometric deep learning model for structure-based protein binding site prediction. Nat. Methods 2022, 19, 730–739. [Google Scholar] [CrossRef]
- Thieker, D.F.; Maguire, J.B.; Kudlacek, S.T.; Leaver-Fay, A.; Lyskov, S.; Kuhlman, B. Stabilizing proteins, simplified: A Rosetta-based webtool for predicting favorable mutations. Protein Sci. 2022, 31, e4428. [Google Scholar] [CrossRef] [PubMed]
- Ferla, M.P.; Pagnamenta, A.T.; Koukouflis, L.; Taylor, J.C.; Marsden, B.D. Venus: Elucidating the impact of amino acid variants on protein function beyond structure destabilisation. J. Mol. Biol. 2022, 434, 167567. [Google Scholar] [CrossRef]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef]
- Chen, R.; Braun, G.B.; Luo, X.; Sugahara, K.N.; Teesalu, T.; Ruoslahti, E. Application of a proapoptotic peptide to intratumorally spreading cancer therapy. Cancer Res. 2013, 73, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.-K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Possemato, R.; Marks, K.M.; Shaul, Y.D.; Pacold, M.E.; Kim, D.; Birsoy, K.; Sethumadhavan, S.; Woo, H.-K.; Jang, H.G.; Jha, A.K.; et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011, 476, 346–350. [Google Scholar] [CrossRef] [PubMed]


| Mutation Type | Missense Mutations | Nonsense Mutations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PTRH2 variant a | c.254A > C | c.68 T > C | c.280 T > A | c.269_270delCT | c.324G > A | c.127dupA | c.328G > T | |||||
| PTRH2 protein variant | p.Gln85Pro (Q85P) | p.Val23Ala (V23A) | p.Tyr94Asn (Y94N) | p.Ala90Glyfs*13 (A90Gfs*13) | p.Trp108* (W108*) | p.Ser43Lysfs*11 (S43Kfs*11) | p.Glu110* (E110*) | |||||
| Reference | [23] | [2] | [3] | [24] | Current Case | [25] | [20] | [1] | [4] | [5] | [26] | [6] |
| Ethnicity | Arab | Arab | Arab | Arab | Arab | Iranian | Japanese | Turkish | Arab | Indian | NR b | Indian |
| Number of patients | 1 | 5 | 3 | 4 | 1 | 1 | 1 | 2 | 3 | 1 | 2 | 1 |
| Clinical Features | ||||||||||||
| Motor delay | 14/14 | 1/1 | 0/1 | 2/2 | 3/3 | 0/1 | 2/2 | 1/1 | ||||
| Intellectual disability | 11/14 | 0/1 | 1/1 | 2/2 | 3/3 | 1/1 | 2/2 | 1/1 | ||||
| Hearing impairment | 10/14 | 0/1 | 1/1 | 2/2 | 1/3 | 1/1 | 2/2 | 1/1 | ||||
| Deformity of head and face | 10/14 | 0/1 | 1/1 | 2/2 | 2/3 | 0/1 | NR | 0/1 | ||||
| Hand deformity | 10/14 | 0/1 | 1/1 | 2/2 | 1/3 | 0/1 | 2/2 | 0/1 | ||||
| Distal weakness | 11/13 c | 1/1 | 1/1 | 2/2 | 2/3 | 0/1 | NR | 1/1 | ||||
| Ataxia | 6/9 | NR | 1/1 | 2/2 | 3/3 | 0/1 | 2/2 | 1/1 | ||||
| Cerebellar atrophy/hypo-plasia | 2/9 | NR | 1/1 | 2/2 | 2/3 | 0/1 | NR | 1/1 | ||||
| Neuropathy | 8/9 | 0/1 | 1/1 | 2/2 | 3/3 | 1/1 | 2/2 | 1/1 | ||||
| Liver abnormality | 1/9 | 0/1 | 0/1 | 2/2 | 0/3 | 0/1 | NR | 1/1 | ||||
| Pancreatic abnormality | 1/9 | 0/1 | 1/1 | 1/2 | 2/3 | 0/1 | 1/2 | 1/1 | ||||
| Hypothyroid-ism | 0/14 | 0/1 | 1/1 | 2/2 | 0/3 | 0/1 | 1/2 | 0/1 | ||||
| Diabetes mellitus | 0/13 | NR | 0/1 | 2/2 | 2/3 | 1/1 | 1/2 | 1/1 | ||||
| Factors | Relative Frequency (N = 25) |
|---|---|
| Total number of reported families | 14 |
| Number of patients affected by: | |
| Missense mutations: | 16 (64%) |
| (A) c.254A > C, p.Gln85Pro | 14 (56%) |
| (B) c.68T > C, p.Val23Ala | 1 (4%) |
| (C) c.280T > A, p.Tyr94Asn | 1 (4%) |
| Nonsense mutations: | 9 (36%) |
| (A) Nonsense point mutation (c.324G > A, p.Trp108*, c.328G > T, p.Glu110*) | 4 (16%) |
| (B) Nonsense nucleotide deletion (c.269_270delCT, p.Ala90Glyfs*13) | 2 (8%) |
| (C) Nonsense nucleotide duplication (c.127dupA, p.Ser43Lysfs*11) | 3 (12%) |
| Motor delay | 23/25 (92%) |
| Intellectual disability | 21/25 (84%) |
| Hearing impairment | 20/25 (80%) |
| Deformity of head and face | 16/23 c (69.5%) |
| Hand deformity | 16/25 (64%) |
| Distal weakness | 19/22 (86.4%) |
| Ataxia | 15/19 (79%) |
| Cerebellar atrophy/hypoplasia | 8/17 (47%) |
| Neuropathy | 18/20 (90%) |
| Liver abnormality | 4/18 (22.2%) |
| Pancreatic abnormality | 7/20 (35%) |
| Hypothyroidism | 4/25 (16%) |
| Diabetes mellitus | 7/23 (30.4%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharkia, R.; Jain, S.; Mahajnah, M.; Habib, C.; Azem, A.; Al-Shareef, W.; Zalan, A. PTRH2 Gene Variants: Recent Review of the Phenotypic Features and Their Bioinformatics Analysis. Genes 2023, 14, 1031. https://doi.org/10.3390/genes14051031
Sharkia R, Jain S, Mahajnah M, Habib C, Azem A, Al-Shareef W, Zalan A. PTRH2 Gene Variants: Recent Review of the Phenotypic Features and Their Bioinformatics Analysis. Genes. 2023; 14(5):1031. https://doi.org/10.3390/genes14051031
Chicago/Turabian StyleSharkia, Rajech, Sahil Jain, Muhammad Mahajnah, Clair Habib, Abdussalam Azem, Wasif Al-Shareef, and Abdelnaser Zalan. 2023. "PTRH2 Gene Variants: Recent Review of the Phenotypic Features and Their Bioinformatics Analysis" Genes 14, no. 5: 1031. https://doi.org/10.3390/genes14051031
APA StyleSharkia, R., Jain, S., Mahajnah, M., Habib, C., Azem, A., Al-Shareef, W., & Zalan, A. (2023). PTRH2 Gene Variants: Recent Review of the Phenotypic Features and Their Bioinformatics Analysis. Genes, 14(5), 1031. https://doi.org/10.3390/genes14051031





