Genome-Wide Association Study Identified Novel SNPs Associated with Chlorophyll Content in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Field Experiments
2.3. Statistical Analysis of Phenotypes
2.4. Genome-Wide Association Mapping
2.5. Prediction of Candidate Genes
2.6. Heat Map of Candidate Genes Expression
2.7. RNA Extraction and RT-qPCR
3. Results
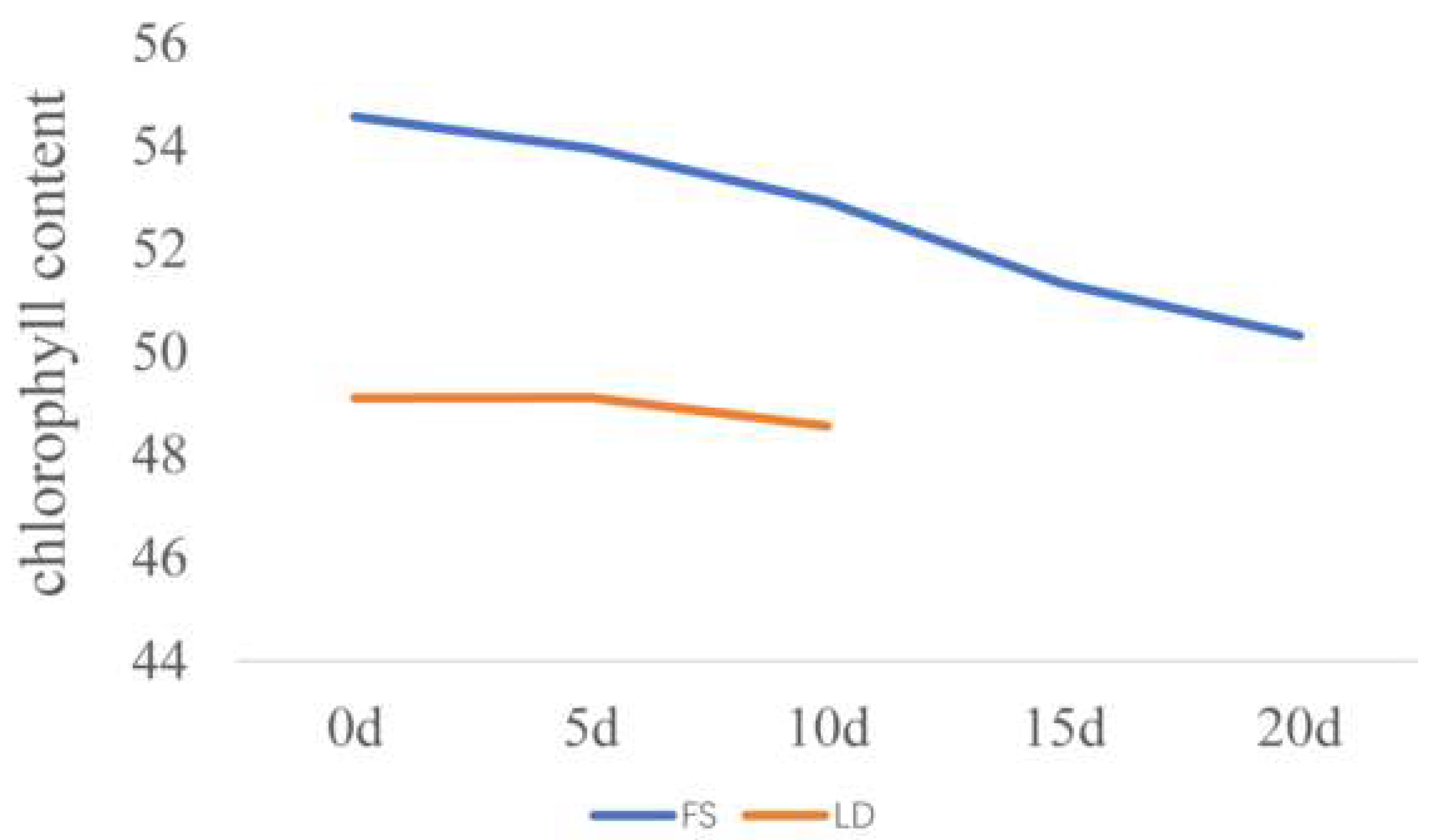
3.1. Chlorophyll Content Diversity and Heritability at Silking
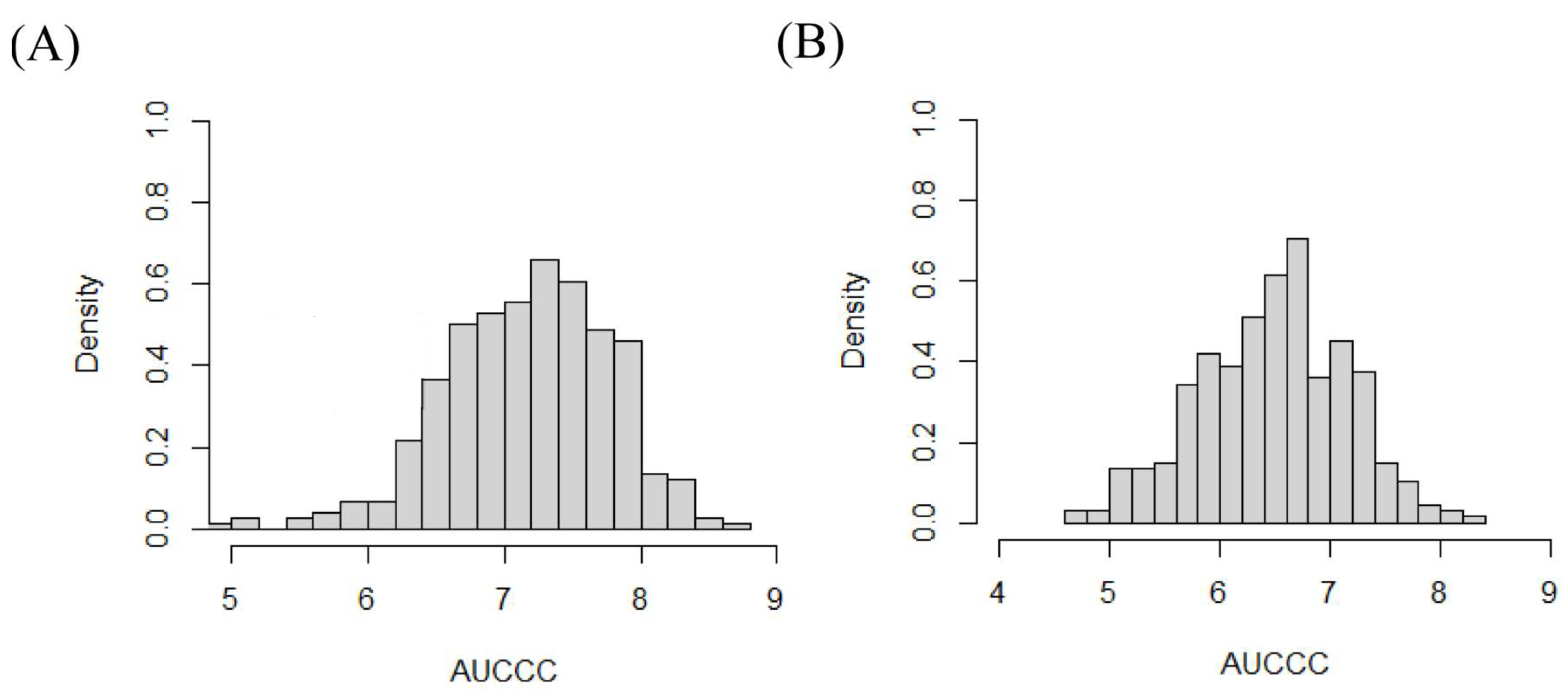
3.2. AUCCC and Heritability after Silking
3.3. Correlations of Chlorophyll Content with Other Plant Developmental Processes
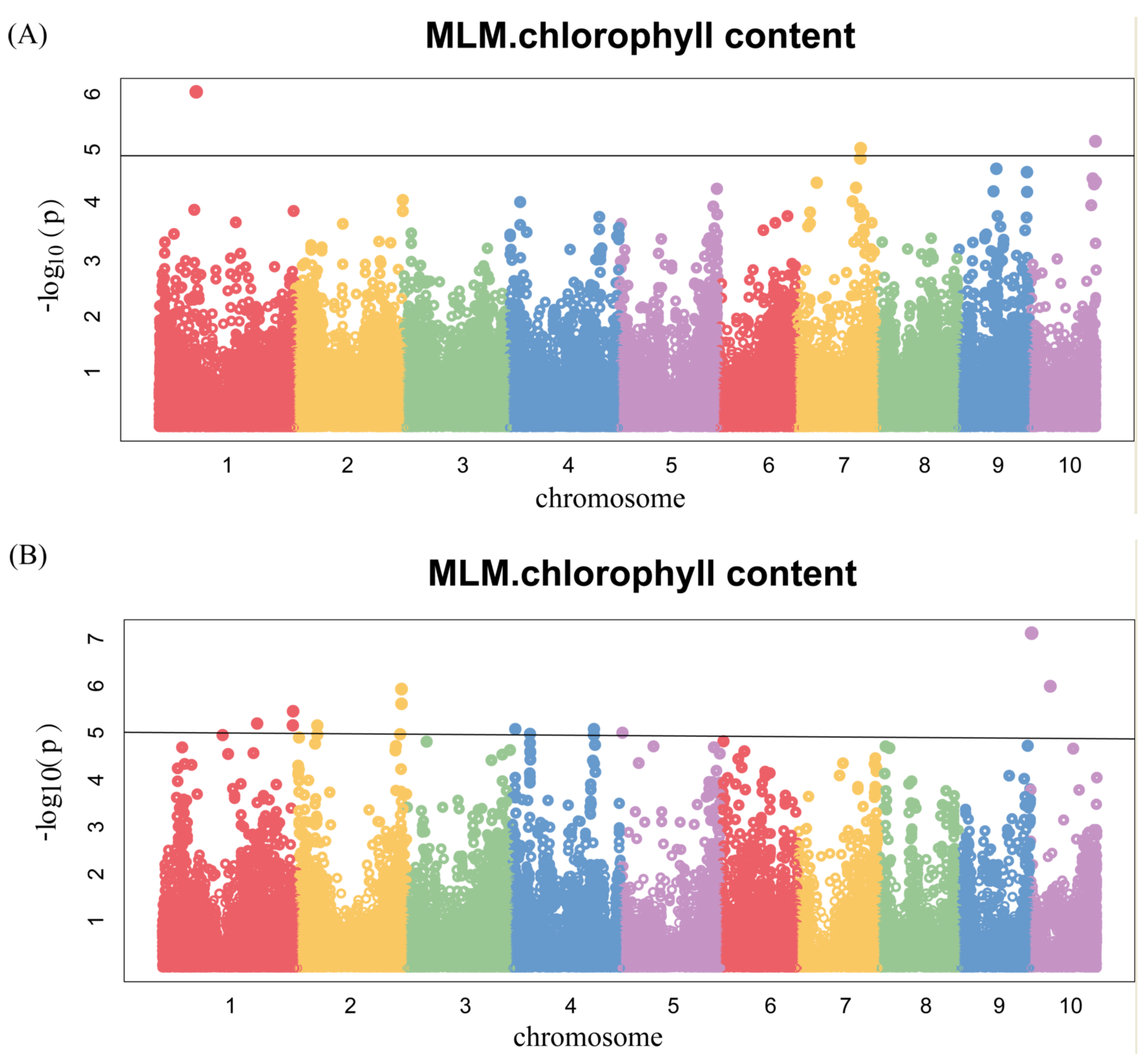
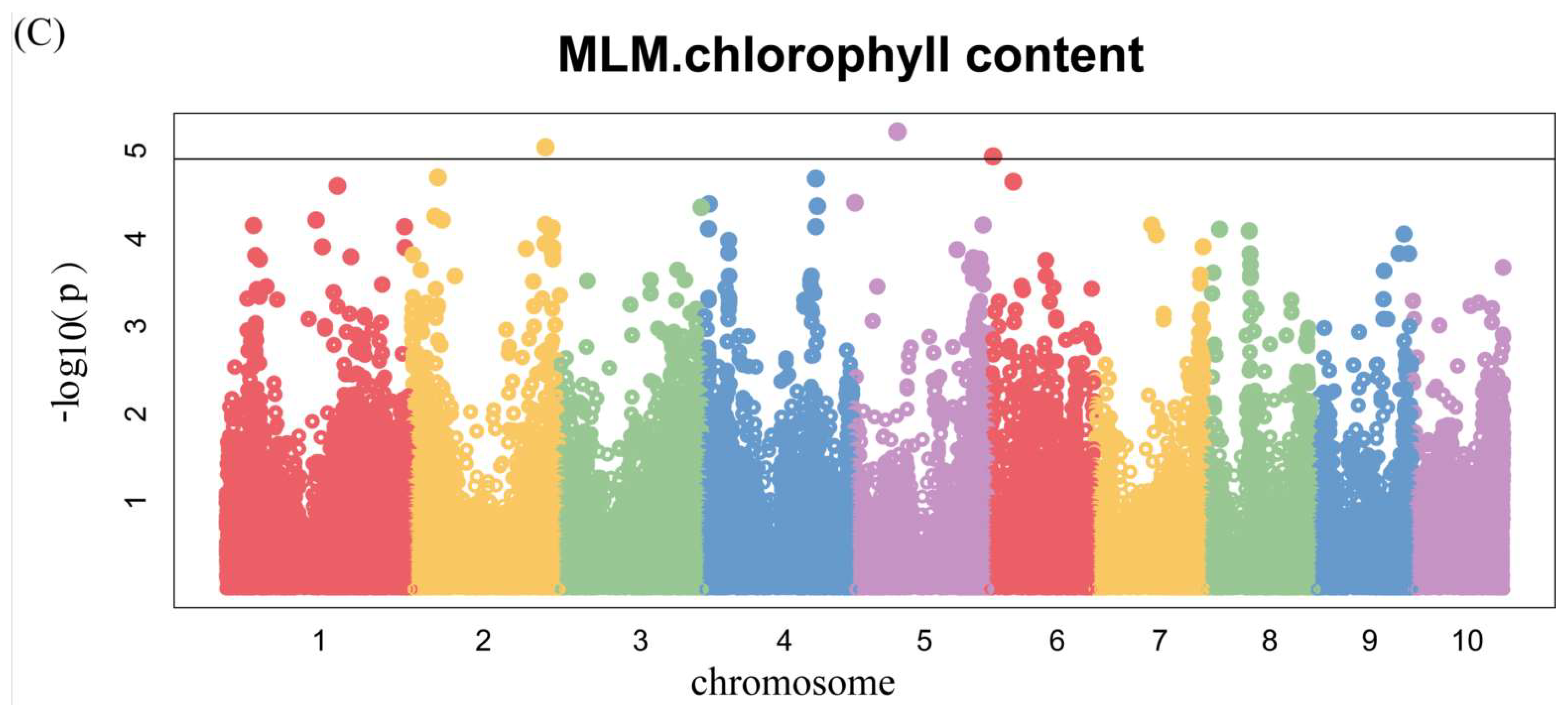
3.4. Genome-Wide Association Analysis
3.5. Candidate Genes
3.6. Expression Pattern of Candidate Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Raines, C.A. Increasing Photosynthetic Carbon Assimilation in C3 Plants to Improve Crop Yield: Current and Future Strategies. Plant Physiol. 2011, 155, 7. [Google Scholar] [CrossRef] [PubMed]
- Grossman, A.R.; Bhaya, D.; Apt, K.E.; Kehoe, D.M. Light-Harvesting Complexes in Oxygenic Photosynthesis: Diversity, Control, and Evolution. Annu. Rev. Genet. 1995, 29, 231–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xie, W.; Xing, H.; Yan, J.; Meng, X.; Li, X.; Fu, X.; Xu, J.; Lian, X.; Yu, S.; et al. Genetic Architecture of Natural Variation in Rice Chlorophyll Content Revealed by a Genome-Wide Association Study. Mol. Plant 2015, 8, 946–957. [Google Scholar] [CrossRef]
- Peng, S.; Khush, G.S.; Virk, P.; Tang, Q.; Zou, Y. Progress in Ideotype Breeding to Increase Rice Yield Potential. Field Crops Res. 2008, 108, 32–38. [Google Scholar] [CrossRef]
- Teng, S.; Qian, Q.; Zeng, D.; Kunihiro, Y.; Fujimoto, K.; Huang, D.; Zhu, L. QTL Analysis of Leaf Photosynthetic Rate and Related Physiological Traits in Rice (Oryza sativa L.). Euphytica 2004, 135, 1–7. [Google Scholar] [CrossRef]
- Li, T.; Yang, H.; Lu, Y.; Dong, Q.; Liu, G.; Chen, F.; Zhou, Y. Comparative Transcriptome Analysis of Differentially Expressed Genes Related to the Physiological Changes of Yellow-Green Leaf Mutant of Maize. PeerJ 2021, 9, e10567. [Google Scholar] [CrossRef]
- Takai, T.; Kondo, M.; Yano, M.; Yamamoto, T. A Quantitative Trait Locus for Chlorophyll Content and Its Association with Leaf Photosynthesis in Rice. Rice 2010, 3, 172–180. [Google Scholar] [CrossRef]
- Wang, L.; Conteh, B.; Fang, L.; Xia, Q.; Nian, H. QTL Mapping for Soybean (Glycine max L.) Leaf Chlorophyll-Content Traits in a Genotyped RIL Population by Using RAD-Seq Based High-Density Linkage Map. BMC Genom. 2020, 21, 739. [Google Scholar] [CrossRef]
- Wang, P.; Grimm, B. Connecting Chlorophyll Metabolism with Accumulation of the Photosynthetic Apparatus. Trends Plant Sci. 2021, 26, 484–495. [Google Scholar] [CrossRef]
- Tanaka, R.; Tanaka, A. Tetrapyrrole Biosynthesis in Higher Plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef]
- Mochizuki, N.; Tanaka, R.; Grimm, B.; Masuda, T.; Moulin, M.; Smith, A.G.; Tanaka, A.; Terry, M.J. The Cell Biology of Tetrapyrroles: A Life and Death Struggle. Trends Plant Sci. 2010, 15, 488–498. [Google Scholar] [CrossRef]
- Tanaka, R.; Tanaka, A. Chlorophyll Cycle Regulates the Construction and Destruction of the Light-Harvesting Complexes. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 968–976. [Google Scholar] [CrossRef]
- Christ, B.; Hörtensteiner, S. Mechanism and Significance of Chlorophyll Breakdown. J. Plant Growth Regul. 2014, 33, 4–20. [Google Scholar] [CrossRef]
- Lin, Y.-P.; Wu, M.-C.; Charng, Y. Identification of a Chlorophyll Dephytylase Involved in Chlorophyll Turnover in Arabidopsis. Plant Cell 2016, 28, 2974–2990. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, X.; Zhang, F.; Xu, Z.; Chen, W.; Li, Y. Identification and Fine Mapping of QCTH4, a Quantitative Trait Loci Controlling the Chlorophyll Content from Tillering to Heading in Rice (Oryza sativa L.). J. Hered. 2012, 103, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.Y.; Bollivar, D.W.; Bauer, C.E. Genetic analysis of chlorophyll biosynthesis. Annu. Rev. Genet. 1997, 31, 61–89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-H.; Li, S.-Y.; Wang, L.; Ye, W.-J.; Zeng, D.-L.; Rao, Y.-C.; Peng, Y.-L.; Hu, J.; Yang, Y.-L.; Xu, J.; et al. LSCHL4 from Japonica Cultivar, Which Is Allelic to NAL1, Increases Yield of Indica Super Rice 93-11. Mol. Plant 2014, 7, 1350–1364. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Hu, S.; Wu, L.; Ge, C.; Cui, Y.; Chen, P.; Xu, J.; Dong, G.; Guo, L.; Qian, Q. Fine Mapping a Major QTL QFCC7 L for Chlorophyll Content in Rice (Oryza sativa L.) Cv. PA64s. Plant Growth Regul. 2017, 81, 81–90. [Google Scholar] [CrossRef]
- Sakai, T.; Abe, A.; Shimizu, M.; Terauchi, R. RIL-StEp: Epistasis Analysis of Rice Recombinant Inbred Lines Reveals Candidate Interacting Genes That Control Seed Hull Color and Leaf Chlorophyll Content. G3 Genes Genomes Genet. 2021, 11, jkab130. [Google Scholar] [CrossRef] [PubMed]
- Salvi, S.; Sponza, G.; Morgante, M.; Tomes, D.; Niu, X.; Fengler, K.A.; Meeley, R.; Ananiev, E.V.; Svitashev, S.; Bruggemann, E.; et al. Conserved Noncoding Genomic Sequences Associated with a Flowering-Time Quantitative Trait Locus in Maize. Proc. Natl. Acad. Sci. USA 2007, 104, 11376–11381. [Google Scholar] [CrossRef]
- Ducrocq, S.; Giauffret, C.; Madur, D.; Combes, V.; Dumas, F.; Jouanne, S.; Coubriche, D.; Jamin, P.; Moreau, L.; Charcosset, A. Fine Mapping and Haplotype Structure Analysis of a Major Flowering Time Quantitative Trait Locus on Maize Chromosome 10. Genetics 2009, 183, 1555–1563. [Google Scholar] [CrossRef] [PubMed]
- Ravelombola, W.S.; Qin, J.; Shi, A.; Nice, L.; Bao, Y.; Lorenz, A.; Orf, J.H.; Young, N.D.; Chen, S. Genome-Wide Association Study and Genomic Selection for Soybean Chlorophyll Content Associated with Soybean Cyst Nematode Tolerance. BMC Genomics 2019, 20, 904. [Google Scholar] [CrossRef]
- Dhanapal, A.P.; Ray, J.D.; Singh, S.K.; Hoyos-Villegas, V.; Smith, J.R.; Purcell, L.C.; Fritschi, F.B. Genome-Wide Association Mapping of Soybean Chlorophyll Traits Based on Canopy Spectral Reflectance and Leaf Extracts. BMC Plant Biol. 2016, 16, 174. [Google Scholar] [CrossRef]
- Herritt, M.; Dhanapal, A.P.; Purcell, L.C.; Fritschi, F.B. Identification of Genomic Loci Associated with 21chlorophyll Fluorescence Phenotypes by Genome-Wide Association Analysis in Soybean. BMC Plant Biol. 2018, 18, 312. [Google Scholar] [CrossRef]
- Teng, S.; Wang, H.; Liang, H.; Xin, H.; Li, S.; Lang, Z. Genome-wide Association Study of Chlorophyll Content in Maize Leaves. Biotechnol. Bull. 2017, 33, 98–107. (In Chinese) [Google Scholar] [CrossRef]
- Yang, W.; Guo, Z.; Huang, C.; Duan, L.; Chen, G.; Jiang, N.; Fang, W.; Feng, H.; Xie, W.; Lian, X.; et al. Combining High-Throughput Phenotyping and Genome-Wide Association Studies to Reveal Natural Genetic Variation in Rice. Nat. Commun. 2014, 5, 5087. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic Variation in ZmVPP1 Contributes to Drought Tolerance in Maize Seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef]
- Sun, D.; Chen, S.; Cui, Z.; Lin, J.; Liu, M.; Jin, Y.; Zhang, A.; Gao, Y.; Cao, H.; Ruan, Y. Genome-Wide Association Study Reveals the Genetic Basis of Brace Root Angle and Diameter in Maize. Front. Genet. 2022, 13, 963852. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liang, Y.; Liu, M.; Zhang, Q.; Wang, Z.; Fan, J.; Ruan, Y.; Zhang, A.; Dong, X.; Yue, J.; et al. Genome-Wide Association Studies Provide Insights Into the Genetic Architecture of Seed Germination Traits in Maize. Front. Plant Sci. 2022, 13, 930438. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Carena, M.J.; Uphaus, J. Area Under the Dry Down Curve (AUDDC): A Method to Evaluate Rate of Dry Down in Maize. Crop Sci. 2010, 50, 2347–2354. [Google Scholar] [CrossRef]
- Knapp, S.J. Confidence Intervals for Heritability for Two-Factor Mating Design Single Environment Linear Models. Theoret. Appl. Genet. 1986, 72, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-Wide Efficient Mixed-Model Analysis for Association Studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Cui, Z.; Luo, J.; Qi, C.; Ruan, Y.; Li, J.; Zhang, A.; Yang, X.; He, Y. Genome-Wide Association Study (GWAS) Reveals the Genetic Architecture of Four Husk Traits in Maize. BMC Genomics 2016, 17, 946. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, H.; Ni, P.; Yu, S.; Dong, H.; Zhang, A.; Cao, H.; Zhang, L.; Ruan, Y.; Cui, Z. Genome-Wide Association Study Dissects the Genetic Architecture of Maize Husk Tightness. Front. Plant Sci. 2020, 11, 861. [Google Scholar] [CrossRef]
- Nehe, A.S. Nitrogen Partitioning and Remobilization in Relation to Leaf Senescence, Grain Yield and Protein Concentration in Indian Wheat Cultivars. Field Crops Res. 2020, 251, 107778. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Guyer, L.; Hörtensteiner, S. Chlorophyll and Chlorophyll Catabolite Analysis by HPLC. In Plant Senescence; Guo, Y., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1744, pp. 223–235. ISBN 978-1-4939-7670-6. [Google Scholar]
- Masojć, P.; Milczarski, P.; Kruszona, P. Comparative Analysis of Genetic Architectures for Nine Developmental Traits of Rye. J. Appl. Genetics 2017, 58, 297–305. [Google Scholar] [CrossRef]
- Borrego-Benjumea, A.; Carter, A.; Zhu, M.; Tucker, J.R.; Zhou, M.; Badea, A. Genome-Wide Association Study of Waterlogging Tolerance in Barley (Hordeum vulgare L.) Under Controlled Field Conditions. Front. Plant Sci. 2021, 12, 711654. [Google Scholar] [CrossRef]
- Young, G.B.; Jack, D.L.; Smith, D.W.; Saier, M.H., Jr. The Amino Acid/Auxin:Proton Symport Permease Family. Biochim. Et Biophys. Acta 1999, 1415, 306–322. [Google Scholar] [CrossRef]
- Zhang, L.; Garneau, M.G.; Majumdar, R.; Grant, J.; Tegeder, M. Improvement of Pea Biomass and Seed Productivity by Simultaneous Increase of Phloem and Embryo Loading with Amino Acids. Plant J. 2015, 81, 134–146. [Google Scholar] [CrossRef]
- Perchlik, M.; Tegeder, M. Leaf Amino Acid Supply Affects Photosynthetic and Plant Nitrogen Use Efficiency under Nitrogen Stress. Plant Physiol. 2018, 178, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, B.; Chen, J.; Wang, X.; Wang, R.; Peng, S.; Chen, L.; Ma, L.; Luo, J. Identification of Rubisco RbcL and RbcS in Camellia oleifera and Their Potential as Molecular Markers for Selection of High Tea Oil Cultivars. Front. Plant Sci. 2015, 6, 189. [Google Scholar] [CrossRef]
- Yadav, S.; Mishra, A.; Jha, B. Elevated CO2 Leads to Carbon Sequestration by Modulating C4 Photosynthesis Pathway Enzyme (PPDK) in Suaeda Monoica and S. Fruticosa. J. Photochem. Photobiol. B Biol. 2018, 178, 310–315. [Google Scholar] [CrossRef]
- Azoulay-Shemer, T.; Palomares, A.; Bagheri, A.; Israelsson-Nordstrom, M.; Engineer, C.B.; Bargmann, B.O.R.; Stephan, A.B.; Schroeder, J.I. Guard Cell Photosynthesis Is Critical for Stomatal Turgor Production, yet Does Not Directly Mediate CO 2- and ABA-induced Stomatal Closing. Plant J. 2015, 83, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, S.; Cao, B.; Cao, D.; Leng, G.; Li, H.; Yin, L.; Shan, L.; Deng, X. Genotypic Variation in Growth and Physiological Response to Drought Stress and Re-Watering Reveals the Critical Role of Recovery in Drought Adaptation in Maize Seedlings. Front. Plant Sci. 2016, 6, 1241. [Google Scholar] [CrossRef]
- Singh, R.; Parihar, P.; Prasad, S.M. Sulfur and Calcium Simultaneously Regulate Photosynthetic Performance and Nitrogen Metabolism Status in As-Challenged Brassica juncea L. Seedlings. Front. Plant Sci. 2018, 9, 772. [Google Scholar] [CrossRef]
- Song, Y.; Li, C.; Zhu, Y.; Guo, P.; Wang, Q.; Zhang, L.; Wang, Z.; Di, H. Overexpression of ZmIPT2 Gene Delays Leaf Senescence and Improves Grain Yield in Maize. Front. Plant Sci. 2022, 13, 963873. [Google Scholar] [CrossRef]
- Xue, Y.; Dong, H.; Huang, H.; Li, S.; Shan, X.; Li, H.; Liu, H.; Xia, D.; Su, S.; Yuan, Y. Mutation in Mg-Protoporphyrin IX Monomethyl Ester (Oxidative) Cyclase Gene ZmCRD1 Causes Chlorophyll-Deficiency in Maize. Front. Plant Sci. 2022, 13, 912215. [Google Scholar] [CrossRef] [PubMed]
- Yamatani, H.; Ito, T.; Nishimura, K.; Yamada, T.; Sakamoto, W.; Kusaba, M. Genetic Analysis of Chlorophyll Synthesis and Degradation Regulated by balance of chlorophyll metabolism. Plant Physiol. 2022, 189, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Myles, S.; Peiffer, J.; Brown, P.J.; Ersoz, E.S.; Zhang, Z.; Costich, D.E.; Buckler, E.S. Association Mapping: Critical Considerations Shift from Genotyping to Experimental Design. Plant Cell 2009, 21, 2194–2202. [Google Scholar] [CrossRef] [PubMed]
- Miculan, M.; Nelissen, H.; Hassen, M.B.; Marroni, F.; Inze, D.; Enrico, M.; Dell’Acqua, M. A Forward Genetics Approach Integrating Genome-Wide Association Study and Expression Quantitative Trait Locus Mapping to Dissect Leaf Development in Maize (Zea mays). Plant J. 2021, 107, 1056–1071. [Google Scholar] [CrossRef]
- Huang, C.; Nie, X.; Shen, C.; You, C.; Li, W.; Zhao, W.; Zhang, X.; Lin, Z. Population Structure and Genetic Basis of the Agronomic Traits of Upland Cotton in China Revealed by a Genome-Wide Association Study Using High-Density SNPs. Plant Biotechnol. J. 2017, 15, 1374–1386. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Niu, X.; Yang, Y.; Wang, S.; Xu, Q.; Yuan, X.; Yu, H.; Wang, Y.; Wang, S.; Feng, Y.; et al. Divergent Hd1, Ghd7, and DTH7 Alleles Control Heading Date and Yield Potential of Japonica Rice in Northeast China. Front. Plant Sci. 2018, 9, 35. [Google Scholar] [CrossRef]
- Jiang, D.; Zhong, S.; McPeek, M.S. Retrospective Binary-Trait Association Test Elucidates Genetic Architecture of Crohn Disease. Am. J. Hum. Genet. 2016, 98, 243–255. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A Unified Mixed-Model Method for Association Mapping that Accounts for Multiple Levels of Relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Yang, J.; Schnable, J.C. Optimising the Identification of Causal Variants across Varying Genetic Architectures in Crops. Plant Biotechnol. J. 2019, 17, 893–905. [Google Scholar] [CrossRef]
- Qu, Z.; Wu, Y.; Hu, D.; Li, T.; Liang, H.; Ye, F.; Xue, J.; Xu, S. Genome-Wide Association Analysis for Candidate Genes Contributing to Kernel-Related Traits in Maize. Front. Plant Sci. 2022, 13, 872292. [Google Scholar] [CrossRef] [PubMed]
- Rashid, Z.; Kaur, H.; Babu, V.; Singh, P.K.; Harlapur, S.I.; Nair, S.K. Identification and Validation of Genomic Regions Associated with Charcoal Rot Resistance in Tropical Maize by Genome-Wide Association and Linkage Mapping. Front. Plant Sci. 2021, 12, 726767. [Google Scholar] [CrossRef]
- Yi, M.; Li, A.; Zhou, L.; Chu, Q.; Song, Y.; Wu, K. The Global Burden and Attributable Risk Factor Analysis of Acute Myeloid Leukemia in 195 Countries and Territories from 1990 to 2017: Estimates Based on the Global Burden of Disease Study 2017. J. Hematol. Oncol. 2020, 13, 72. [Google Scholar] [CrossRef]
- Paraschivu, M.; Cotuna, O.; Paraschivu, M. The use of the area under the disease progress curve (AUDPC) to assess the epidemics of Septoria tritici in winter wheat. Res. J. Agric. Sci. 2013, 45, 193–201. [Google Scholar]
- Irfaq, M.; Ajab, M.; Khattak, G.S.S.; Mohammad, T.; Shah, S.J.A. Genetic Behavior of Controlling Area Under Disease Progress Curve for Stripe Rust (Puccinia striiformis f. Sp. Tritici ) in Two Wheat (Triticum aestivum) Crosses. Phytopathology 2009, 99, 1265–1272. [Google Scholar] [CrossRef]
- Shao, G.; Li, Z.; Ning, T.; Zheng, Y. Responses of Photosynthesis, Chlorophyll Fluorescence, and Grain Yield of Maize to Controlledrelease Urea and Irrigation after Anthesis. J. Plant Nutr. Soil Sci. 2013, 176, 8. [Google Scholar] [CrossRef]
- Chang, C.; Lu, J.; Zhang, H.-P.; Ma, C.-X.; Sun, G. Copy Number Variation of Cytokinin Oxidase Gene Tackx4 Associated with Grain Weight and Chlorophyll Content of Flag Leaf in Common Wheat. PLoS ONE 2015, 10, e0145970. [Google Scholar] [CrossRef]
- Ramesh, K.; Chandrasekaran, B.; Balasubramanian, T.N.; Bangarusamy, U.; Sivasamy, R.; Sankaran, N. Chlorophyll Dynamics in Rice (Oryza sativa) Before and After Flowering Based on SPAD (Chlorophyll) Meter Monitoring and Its Relation with Grain Yield. J. Agron. Crop Sci. 2002, 188, 4. [Google Scholar] [CrossRef]
- Wang, F.; Wang, G.; Li, X.; Huang, J.; Zheng, J. Heredity, Physiology and Mapping of a Chlorophyll Content Gene of Rice (Oryza sativa L.). J. Plant Physiol. 2008, 165, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, H.; Bo, C.; Dai, W.; Zhang, X.; Cai, R.; Gu, L.; Ma, Q.; Jiang, H.; Zhu, J.; et al. Genome-Wide Association Study of Maize Plant Architecture Using F1 Populations. Plant Mol. Biol. 2019, 99, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Geshnizjani, N.; Snoek, B.L.; Willems, L.A.J.; Rienstra, J.A.; Nijveen, H.; Hilhorst, H.W.M.; Ligterink, W. Detection of QTLs for Genotype × Environment Interactions in Tomato Seeds and Seedlings. Plant Cell Environ. 2020, 43, 1973–1988. [Google Scholar] [CrossRef]
- Brumme, S.; Kruft, V.; Schmitz, U.K.; Braun, H.-P. New Insights into the Co-Evolution of Cytochrome C Reductase and the Mitochondrial Processing Peptidase. J. Biol. Chem. 1998, 273, 13143–13149. [Google Scholar] [CrossRef]
- Ow, Y.-L.P.; Green, D.R.; Hao, Z.; Mak, T.W. Cytochrome c: Functions beyond Respiration. Nat. Rev. Mol. Cell Biol. 2008, 9, 532–542. [Google Scholar] [CrossRef]
- Martínez-Fábregas, J.; Díaz-Moreno, I.; González-Arzola, K.; Janocha, S.; Navarro, J.A.; Hervás, M.; Bernhardt, R.; Díaz-Quintana, A.; De la Rosa, M.Á. New Arabidopsis thaliana Cytochrome c Partners: A Look Into the Elusive Role of Cytochrome c in Programmed Cell Death in Plants. Mol. Cell. Proteom. 2013, 12, 3666–3676. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.; Di, C.; Zhang, Q.; Zhang, K.; Wang, C.; You, Q.; Yan, H.; Dai, S.Y.; Yuan, J.S.; et al. JAZ7 Negatively Regulates Dark-Induced Leaf Senescence in Arabidopsis. J. Exp. Bot. 2016, 67, 751–762. [Google Scholar] [CrossRef]
- Wang, H.-L.; Zhang, Y.; Wang, T.; Yang, Q.; Yang, Y.; Li, Z.; Li, B.; Wen, X.; Li, W.; Yin, W.; et al. An Alternative Splicing Variant of PtRD26 Delays Leaf Senescence by Regulating Multiple NAC Transcription Factors in Populus. Plant Cell 2021, 33, 1594–1614. [Google Scholar] [CrossRef]
- Cernac, A.; Benning, C. WRINKLED1 Encodes an AP2/EREB Domain Protein Involved in the Control of Storage Compound Biosynthesis in Arabidopsis: WRI1 Controls Seed Oil Biosynthesis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP Transcription Factors Are Part of Gene Regulatory Networks and Integrate Metabolic, Hormonal and Environmental Signals in Stress Acclimation and Retrograde Signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xia, H.; Li, X.-J.; Hu, R.; Chen, Y.; Li, X.-B. Overexpression of a Cotton Gene That Encodes a Putative Transcription Factor of AP2/EREBP Family in Arabidopsis Affects Growth and Development of Transgenic Plants. PLoS ONE 2013, 8, e78635. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, T. Expansion and Stress Responses of the AP2/EREBP Superfamily in Cotton. BMC Genomics 2017, 18, 118. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, T.E.; Liu, J.; Li, Q.; Xue, H.; Wang, G.; Li, L.; Fontana, J.E.; Davis, K.E.; Liu, W.; Zhang, B.; et al. Potassium Deficiency Significantly Affected Plant Growth and Development as Well as MicroRNA-Mediated Mechanism in Wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 1219. [Google Scholar] [CrossRef]
- Sun, J.; Yang, L.; Yang, X.; Wei, J.; Li, L.; Guo, E.; Kong, Y. Using Spectral Reflectance to Estimate the Leaf Chlorophyll Content of Maize Inoculated with Arbuscular Mycorrhizal Fungi Under Water Stress. Front. Plant Sci. 2021, 12, 646173. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Han, B.; Liu, A.; Xu, W. Integrated Lipidomic and Transcriptomic Analysis Reveals Triacylglycerol Accumulation in Castor Bean Seedlings under Heat Stress. Ind. Crops Prod. 2022, 180, 114702. [Google Scholar] [CrossRef]
- Eklund, D.M.; Edqvist, J. Localization of Nonspecific Lipid Transfer Proteins Correlate with Programmed Cell Death Responses during Endosperm Degradation in Euphorbia lagascae Seedlings. Plant Physiol. 2003, 132, 1249–1259. [Google Scholar] [CrossRef]
- Wu, J.; Sun, Y.; Zhao, Y.; Zhang, J.; Luo, L.; Li, M.; Wang, J.; Yu, H.; Liu, G.; Yang, L.; et al. Deficient Plastidic Fatty Acid Synthesis Triggers Cell Death by Modulating Mitochondrial Reactive Oxygen Species. Cell Res. 2015, 25, 621–633. [Google Scholar] [CrossRef]
- Wagoner, J.A.; Sun, T.; Lin, L.; Hanson, M.R. Cytidine Deaminase Motifs within the DYW Domain of Two Pentatricopeptide Repeat-Containing Proteins Are Required for Site-Specific Chloroplast RNA Editing. J. Biol. Chem. 2015, 290, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, J.; Li, Y.; Su, B.; Xu, H.; Shan, X.; Song, C.; Xie, J.; Li, R. PDM3, a Pentatricopeptide Repeat-Containing Protein, Affects Chloroplast Development. J. Exp. Bot. 2017, 68, 5615–5627. [Google Scholar] [CrossRef] [PubMed]







| Source of Variation a | Mean Square | Significance b | H2 c | |
|---|---|---|---|---|
| chlorophyll content | Environment (E) | 10058.20165 | <0.01 ** | 0.67 |
| Genotype (G) | 190.14580 | <0.01 ** | ||
| G × E | 155.13709 | 0.4034 | ||
| AUCCC | Environment (E) | 149.3227937 | <0.01 ** | 0.66 |
| Genotype (G) | 1.1585265 | <0.01 ** | ||
| G × E | 0.405332 | 0.0987 |
| Trait | SNP | Chr | Position (bp) | p Value | Gene | Gene Interval (bp) | Annotation | Pathway |
|---|---|---|---|---|---|---|---|---|
| 17BLUP chlorophyll content | Marker.247949 | 2 | 2.20 × 108 | 9.10 × 10−6 | Zm00001d007009 | Chr2:220125041-220148355 | DNAJ heat shock N-terminal domain-containing protein | Chloroplast targeting, photosystem II repair |
| Zm00001d007011 | Chr2:220152033-220155206 | ATP synthase | Energy metabolism | |||||
| Zm00001d007012 | Chr2:220170458-220175539 | CHLOROPLAST RNA-BINDING PROTEIN | Chloroplast morphogenesis | |||||
| Zm00001d007016 | Chr2:220204271-220219191 | disease resistance protein RGA2 | Disease-resistant | |||||
| Zm00001d007017 | Chr2:220217348-220221222 | thioredoxin-like protein AAED1 chloroplastic | Electronic circulation and daylighting | |||||
| 17FS chlorophyll content | 2376873-7-G | 10 | 1.48 × 108 | 7.08 × 10−6 | Zm00001d026563 | Chr10:148153479-148157015 | AP2/EREBP transcription factor 40 | Plant growth and development |
| Zm00001d026569 | Chr10:148185247-148190302 | chloroplastic palmitoyl-acyl carrier protein thioesterase | De novo synthesis of fatty acids | |||||
| Zm00001d026568 | Chr10:148184610-148195615 | pentatricopeptide repeat-containing protein | Chloroplast development | |||||
| Zm00001d026573 | Chr10:148201459-148207853 | 5-methylthioribose kinase | Methylthioadenosine (MTA) cycle | |||||
| Zm00001d026574 | Chr10:148208912-148217798 | UDP-D-galacturonate | Homogalacturonan biosynthesis | |||||
| 17LD chlorophyll content | Marker.190636 | 2 | 43,281,907 | 6.97 × 10−6 | Zm00001d003403 | Chr2:43279990-43285413 | aaap10—amino acid/auxin permease10 | Amino acid transportation |
| Zm00001d003404 | Chr2:43284025.43288746 | transmembrane protein | Transmembrane | |||||
| Zm00001d003405 | Chr2:43285551-43289511 | protease inhibitor/seed storage/LTP family protein | Signal transduction | |||||
| Zm00001d003406 | Chr2:43298285-43304459 | actin binding protein | Plant growth and development | |||||
| Marker.346487 | 4 | 8,724,268 | 8.32 × 10−6 | Zm00001d027598 | Chr1:8676135-8681000 | cct101—CO CO-LIKE TIMING OF CAB1 protein domain101) | Transcription factors, floral completion | |
| Zm00001d027599 | Chr1:8699800-8704356 | alkane hydroxylase MAH1 | Cuticular wax biosynthesis | |||||
| Zm00001d027601 | Chr1:8772748-8777355 | behenate ω-hydroxylase | Suberin monomers biosynthesis | |||||
| 2504165-22-G | 5 | 2,775,822 | 9.93 × 10−6 | Zm00001d012982 | Chr5:2770681-2776100 | NAD(P)-binding domain containing protein | Energy metabolism | |
| 17LD AUCCC | Marker.571424 | 6 | 1.73 × 108 | 7.40 × 10−6 | Zm00001d039222 | Chr6:173160699-173165510 | cct40—CO CO-LIKE TIMING OF CAB1 protein domain40) | Transcription factors, floral completion |
| Zm00001d039221 | Chr6:173158717-173163744 | nucleoside diphosphate kinase 4 | Signal transduction | |||||
| 17FS AUCCC | 2376873-7-G | 10 | 1.48 × 108 | 5.82 × 10−6 | Zm00001d026563 | Chr10:148153479-148157015 | ereb40—AP2-EREBP-transcription factor 40 | Plant growth and development |
| Zm00001d026568 | Chr10:148184610-148195615 | pentatricopeptide repeat-containing protein, mitochondrial-like | Chloroplast development | |||||
| Zm00001d026569 | Chr10:148185247-148190302 | palmitoyl-acyl carrier protein thioesterase, chloroplastic | De novo synthesis of fatty acids | |||||
| Zm00001d026573 | Chr10:148201459-148207853 | 5-methylthioribose kinase | Methylthioadenosine (MTA) cycle | |||||
| Zm00001d026574 | Chr10:148208912-148217798 | UDP-D-galacturonate | Homogalacturonan biosynthesis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Li, D.; Liu, M.; Cui, Z.; Sun, D.; Li, C.; Zhang, A.; Cao, H.; Ruan, Y. Genome-Wide Association Study Identified Novel SNPs Associated with Chlorophyll Content in Maize. Genes 2023, 14, 1010. https://doi.org/10.3390/genes14051010
Jin Y, Li D, Liu M, Cui Z, Sun D, Li C, Zhang A, Cao H, Ruan Y. Genome-Wide Association Study Identified Novel SNPs Associated with Chlorophyll Content in Maize. Genes. 2023; 14(5):1010. https://doi.org/10.3390/genes14051010
Chicago/Turabian StyleJin, Yueting, Dan Li, Meiling Liu, Zhenhai Cui, Daqiu Sun, Cong Li, Ao Zhang, Huiying Cao, and Yanye Ruan. 2023. "Genome-Wide Association Study Identified Novel SNPs Associated with Chlorophyll Content in Maize" Genes 14, no. 5: 1010. https://doi.org/10.3390/genes14051010
APA StyleJin, Y., Li, D., Liu, M., Cui, Z., Sun, D., Li, C., Zhang, A., Cao, H., & Ruan, Y. (2023). Genome-Wide Association Study Identified Novel SNPs Associated with Chlorophyll Content in Maize. Genes, 14(5), 1010. https://doi.org/10.3390/genes14051010





