Genome-Wide Identification and Gene Expression Analysis of Sweet Cherry Aquaporins (Prunus avium L.) under Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
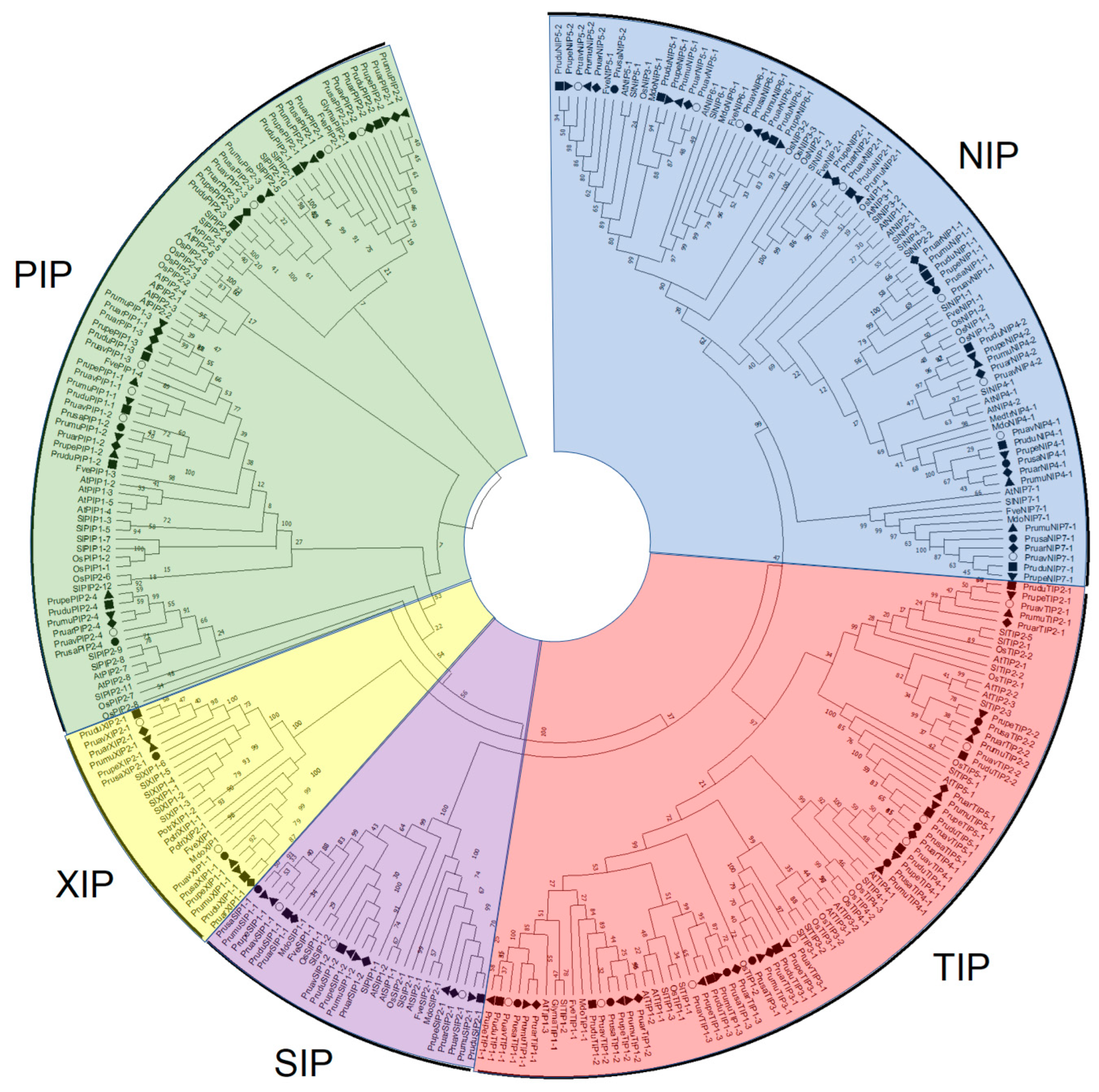
2.1. Identification of Aquaporin Genes and Phylogenetic Analysis
2.2. Microsynteny Relationships of Four Prunus Species
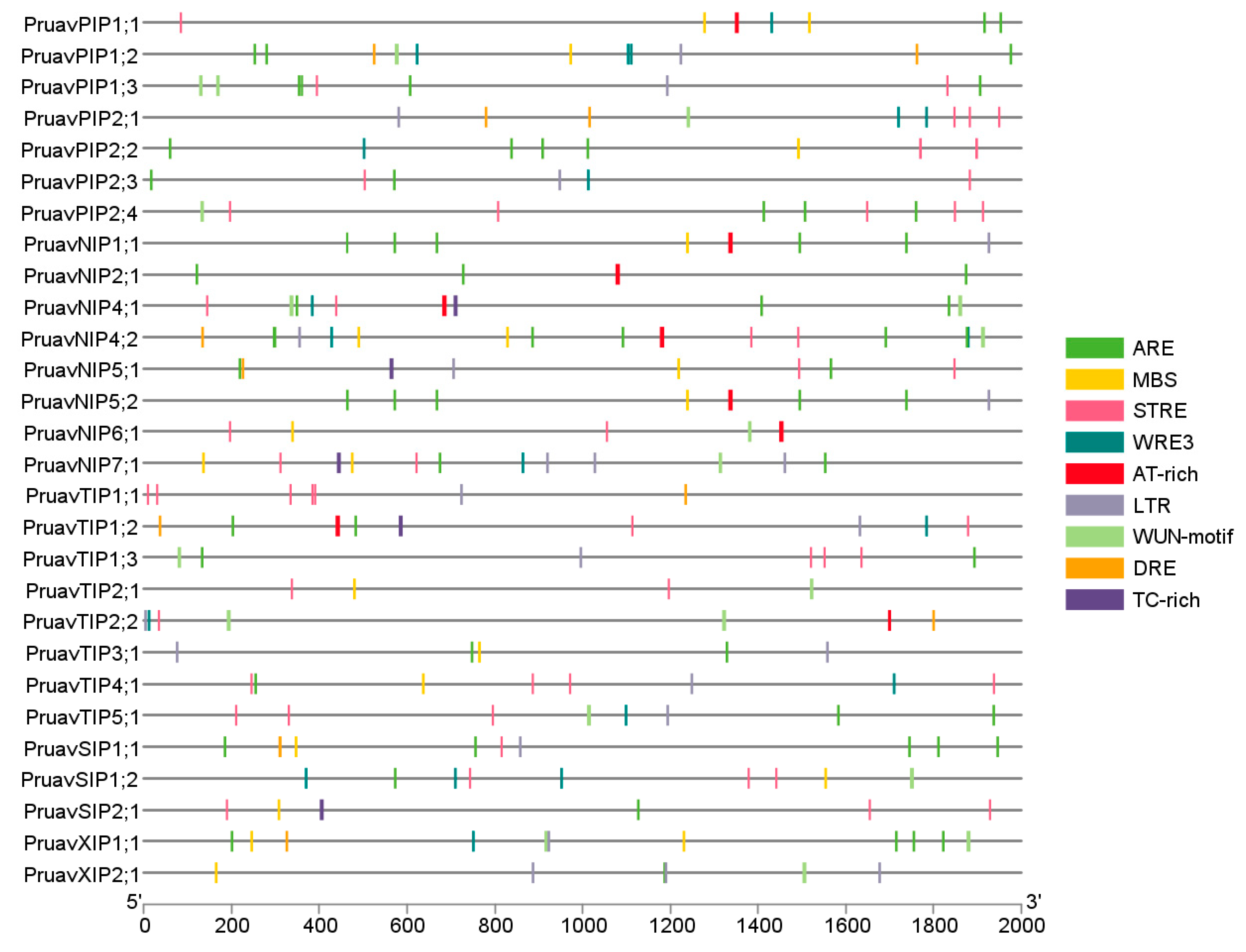
2.3. Cis-Acting Regulatory Element Analysis
2.4. Gene Structure, Characteristic Motif, Physicochemical Features, In Silico Prediction of Transmembrane Domains and Subcellular Localization
2.5. Abiotic Stress Treatments
2.6. RNA Extraction and Transcript Analysis by qRT-PCR
3. Results
3.1. Identification and Analysis of Aquaporin Genes in P. avium
3.2. Analysis of Cis-Acting Regulatory Elements of the Aquaporin Gene Family of P. avium
3.3. Analysis of Exon–Intron Structure
3.4. Structural Characteristics of P. avium Aquaporins
3.4.1. P. avium NIP Subfamily
3.4.2. P. avium PIP Subfamily
3.4.3. P. avium TIP Subfamily
3.4.4. P. avium SIP Subfamily
3.4.5. P. avium XIP Subfamily
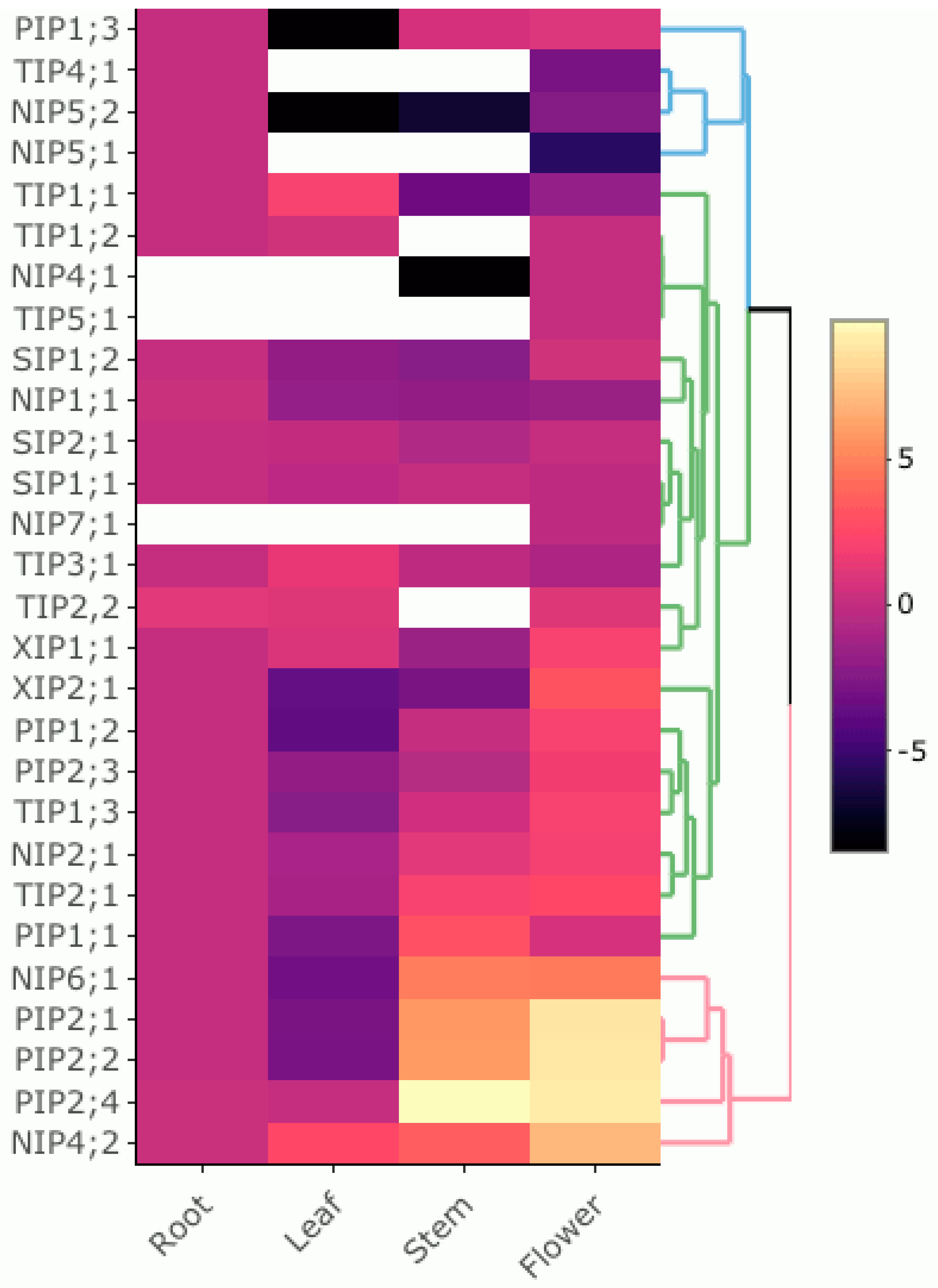
3.5. Expression Pattern of PruavAQP Genes in Different Organs
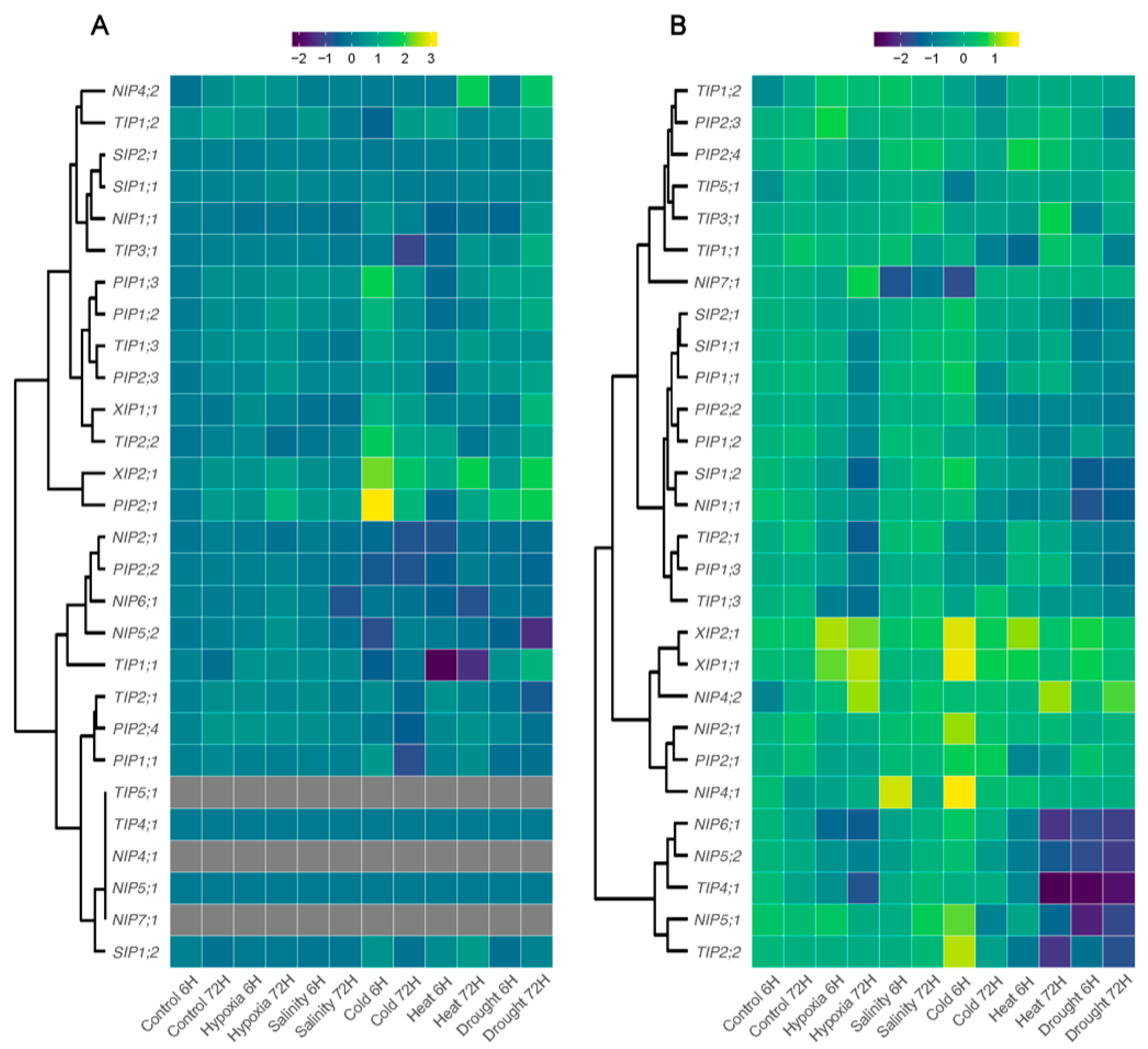
3.6. Expression Profiles of AQPs Genes in Response to Different Abiotic Stresses
4. Discussion
4.1. Identification and In Silico Characterization of the PruavAQP Gene Family
4.2. Organ-Specific Expression
4.3. Expression Profiles of PruavAQPs under Abiotic Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Denker, B.M.; Smith, B.L.; Kuhadja, F.P.; Agre, P. Identification, purification and characterization of a novel Mr 28,000 integral membrane protein from erythrocytes and renal tubules. J. Biol. Chem. 1988, 263, 15634–15642. [Google Scholar] [CrossRef] [PubMed]
- Chaumont, F.; Tyerman, S.D. Aquaporins: Highly regulated channels controlling plant water relations. Plant Physiol. 2014, 164, 1600–1618. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.R.; Shahzad, Z.; Verdoucq, L. Aquaporins in plants. Physiology 2015, 95, 1321–1358. [Google Scholar]
- Zwiazek, J.J.; Xu, H.; Tan, X.; Navarro-Ródenas, A.; Morte, A. Significance of oxygen transport through aquaporins. Sci. Rep. 2017, 7, 40411. [Google Scholar] [CrossRef] [PubMed]
- Behera, B.K.; Parhi, J.; Dehury, B.; Rout, A.K.; Khatei, A.; Devi, A.L.; Mandal, S.C. Molecular characterization and structural dynamics of Aquaporin1 from walking catfish in lipid bilayers. Int. J. Biol. Macromol. 2022, 196, 86–97. [Google Scholar] [CrossRef]
- Walz, T. The three-dimensional structure of aquaporin-1. Nature 1997, 387, 624–627. [Google Scholar] [CrossRef]
- Törnroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; Neutze, R.; Kjellbom, P. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef]
- Froger, A.; Tallur, B.; Thomas, D.; Delamarche, C. Prediction of functional residues in water channels and related proteins. Protein Sci. 1998, 7, 1458–1468. [Google Scholar] [CrossRef]
- Hove, R.; Bhave, M. Plant aquaporins with non-aqua functions: Deciphering the signature sequences. Mol. Biol. 2011, 75, 413–430. [Google Scholar] [CrossRef]
- Mateluna, P.; Salvatierra, A.; Solis, S.; Nuñez, G.; Pimentel, P. Involvement of aquaporin NIP1;1 in the contrasting tolerance response to root hypoxia in Prunus rootstocks. J. Plant Physiol. 2018, 228, 19–28. [Google Scholar] [CrossRef]
- Maurel, C.; Verdoucq, L.; Luu, D.-T.; Santoni, V. Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef]
- Prado, K.; Boursiac, Y.; Tournaire-Roux, C.; Monneuse, J.M.; Postaire, O.; Da Ines, O.; Schäffner, A.R.; Hem, S.; Santoni, V.; Maurel, C. Regulation of Arabidopsis Leaf Hydraulics Involves Light-Dependent Phosphorylation of Aquaporins in Veins. Plant Cell 2013, 25, 1029–1039. [Google Scholar] [CrossRef]
- Sato, R.; Maeshima, M. The ER-localized aquaporin SIP2;1 is involved in pollen germination and pollen tube elongation in Arabidopsis thaliana. Plant Mol. Biol. 2019, 100, 335–349. [Google Scholar] [CrossRef]
- Kurowska, M.M. Aquaporins in cereals-important players in maintaining cell homeostasis under abiotic stress. Genes 2021, 25, 477. [Google Scholar] [CrossRef]
- Su, Y.; Liu, Z.; Sun, J.; Wu, C.; Li, Y.; Zhang, C.; Zhao, L. Genome-wide identification of maize aquaporin and functional analysis during seed germination and seedling establishment. Front. Plant Sci. 2022, 13, 831916. [Google Scholar] [CrossRef]
- Webster, A.D. Rootstock and interstock effects on deciduous fruit tree vigour, precocity, and yield productivity. New Zealand J. Crop Hortic. Sci. 1995, 23, 373–382. [Google Scholar] [CrossRef]
- Warschefsky, E.J.; Klein, L.L.; Frank, M.H.; Chitwood, D.H.; Londo, J.P.; von Wettberg, E.J.B. Rootstocks: Diversity, domestication, and impacts on shoot phenotypes. Trends Plant Sci. 2016, 21, 418–437. [Google Scholar] [CrossRef]
- Schwarz, D. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Sci. Hortic. 2010, 127, 162–171. [Google Scholar] [CrossRef]
- Olden, E.J.; Nybom, N. On the origin of Prunus cerasus L. Hereditas 1968, 59, 327–345. [Google Scholar] [CrossRef]
- Iezzoni, A.; Schmidt, H.; Albertini, A. Cherries (Prunus). Genet. Resour. Temp. Fruit Nut Crops 1991, 290, 111–176. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Calle, A.; Cai, L.; Iezzoni, A.; Wünsch, A. High-density linkage maps constructed in sweet cherry (Prunus avium L.) using cross-and self-pollination populations reveal chromosomal homozygosity in inbred families and non-syntenic regions with the peach genome. Tree Genet. Genomes 2018, 14, 37. [Google Scholar] [CrossRef]
- Baek, S.; Choi, K.; Kim, G.B.; Yu, H.J.; Cho, A.; Jang, H.; Mun, J.H. Draft genome sequence of wild Prunus yedoensis reveals massive inter-specific hybridization between sympatric flowering cherries. Genome Biol. 2018, 19, 127. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Liu, C.; Li, L.; Li, M. Arabidopsis EOD3 homologue PaCYP78A6 affects fruit size and is involved in sweet cherry (Prunus avium L.) fruit ripening. Sci. Hortic. 2019, 246, 57–67. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Zhu, D.; Po, H.; Shizhong, Z.; Shijun, X.; Yue, T.; Xin, C.; Li, X.; Xiaojuan, Z.; et al. Chromosome-scale genome assem. sweet cherry (Prunus avium L.) cv. Tieton obtained using long-read Hi-C sequencing. Hortic Res. 2020, 7, 122. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Murata, K.; Mitsuoka, K.; Hirai, T.; Walz, T.; Agre, P.; Heymann, J.B.; Engel, A.; Fujiyoshi, Y. Structural determinants of water permeation through aquaporin-1. Nature 2000, 407, 599–605. [Google Scholar] [CrossRef]
- Hub, J.S.; de Groot, B.L. Mechanism of selectivity in aquaporins and aquaglyceroporins. Proc. Natl. Acad. Sci. USA 2008, 105, 1198–1203. [Google Scholar] [CrossRef]
- Mitani-Ueno, N.; Yamaji, N.; Zhao, F.J.; Ma, J.F. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 2011, 62, 4391–4398. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Pimentel, P.; Almada, R.D.; Salvatierra, A.; Toro, G.; Arismendi, M.J.; Pino, M.T.; Sagredo, B.; Pinto, M. Physiological and morphological responses of Prunus species with different degree of tolerance to long-term root hypoxia. Sci. Hortic. 2014, 180, 14–23. [Google Scholar] [CrossRef]
- Toro, G.; Pimentel, P.; Salvatierra, A. Effective categorization of tolerance to salt stress through clustering Prunus rootstocks according to their physiological performances. Horticulturae 2021, 7, 542. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Deshmukh, R.A. A precise spacing between the NPA domains of aquaporins is essential for silicon permeability in plants. Plant J. 2015, 83, 489–500. [Google Scholar] [CrossRef]
- Quigley, F.; Rosenberg, J.M.; Shachar-Hill, Y.; Bohnert, H.J. From genome to function: The Arabidopsis aquaporins. Genome Biol. 2001, 3, RESEARCH0001. [Google Scholar] [CrossRef]
- Sakurai, J.; Ishikawa, F.; Yamaguchi, T.; Uemura, M.; Maeshima, M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005, 46, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.B.; Sankararamakrishnan, R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: Characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 2009, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- AR6 Synthesis Report, Climate Change 2023. Available online: https://report.ipcc.ch/ar6syr/pdf/IPCC_AR6_SYR_LongerReport.pdf (accessed on 25 February 2023).
- Heckwolf, M.; Pater, D.; Hanson, D.T.; Kaldenhoff, R. The Arabidopsis thaliana aquaporin AtPIP1;2 is a physiologically relevant CO2 transport facilitator. Plant. J. 2011, 67, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, S.; Lou, Y.; Zhu, C.; Zhao, H.; Li, Y.; Li, X.; Gao, Z. A bamboo leaf-specific aquaporin gene PePIP2;7 is involved in abiotic stress response. Plant Cell Rep. 2021, 40, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Zupin, M.; Sedlar, A.; Kidrič, M.; Meglič, V. Drought-induced expression of aquaporin genes in leaves of two common bean cultivars differing in tolerance to drought stress. J. Plant Res. 2017, 130, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Sahitya, U.L.; Krishna, M.S.R.; Suneetha, P. Integrated approaches to study the drought tolerance mechanism in hot pepper (Capsicum annuum L.). Physiol. Mol. Biol. Plants 2019, 25, 637–647. [Google Scholar] [CrossRef]
- Xue, H.; Zhang, F.; Zhang, Z.H.; Fu, J.F.; Wang, F.; Zhang, B.; Ma, Y. Differences in salt tolerance between diploid and autotetraploid apple seedlings exposed to salt stress. Sci. Hortic. 2015, 190, 24–30. [Google Scholar] [CrossRef]
- Jia, J.; Liang, Y.; Gou, T.; Hu, Y.; Zhu, Y.; Huo, H.; Guo, J.; Gong, H. The expression response of plasma membrane aquaporins to salt stress in tomato plants. Environ. Exp. Bot. 2020, 178, 104190. [Google Scholar] [CrossRef]
- Li, D.D.; Tai, F.J.; Zhang, Z.T.; Li, Y.; Zheng, Y.; Wu, Y.F.; Li, X.B. A cotton gene encodes a tonoplast aquaporin that is involved in cell tolerance to cold stress. Gene 2009, 438, 26–32. [Google Scholar] [CrossRef]
- Ahamed, A.; Murai-Hatano, M.; Ishikawa-Sakurai, J.; Hayashi, H.; Kawamura, Y.; Uemura, M. Cold stress-induced acclimation in rice is mediated by root-specific aquaporins. Plant Cell Physiol. 2012, 53, 1445–1456. [Google Scholar] [CrossRef]
- Wang, Y.; Li, R.; Li, D.; Jia, X.; Zhou, D.; Li, J.; Liu, J. NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 5047–5052. [Google Scholar] [CrossRef]
- Dong, J.; Niu, S.; Qian, J.; Zhou, J.; Zhao, M.; Meng, Y.; Di, B. Identification of Aquaporin Gene Family in Response to Natural Cold Stress in Ligustrum × vicaryi Rehd. Forests 2022, 13, 182. [Google Scholar] [CrossRef]
- Simoes-Araujo, J.L.; Alves-Ferreira, M.; Rumjanek, N.G.; Margis-Pinheiro, M. VuNIP1 (NOD26-like) and VuHSP17. 7 gene expression are regulated in response to heat stress in cowpea nodule. Environ. Exp. Bot. 2008, 63, 256–265. [Google Scholar] [CrossRef]
- He, W.; Liu, M.; Qin, X.; Liang, A.; Chen, Y.; Yin, Y.; Qin, K.; Mu, Z. Genome-wide identification and expression analysis of the aquaporin gene family in Lycium barbarum during fruit ripening and seedling response to heat stress. Curr. Issues Mol. Biol. 2022, 44, 5933–5948. [Google Scholar] [CrossRef]
- Shafqat, W.; Jaskani, M.J.; Maqbool, R.; Chattha, W.S.; Ali, Z.; Naqvi, S.A.; Haider, M.S.; Kha, I.A.; Vincent, C.I. Heat shock protein and aquaporin expression enhance water conserving behavior of citrus under water deficits and high temperature conditions. Environ. Exp. Bot. 2021, 181, 104270. [Google Scholar] [CrossRef]
- Reuscher, S.; Akiyama, M.; Mori, C.; Aoki, K.; Shibata, D.; Shiratake, K. Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 2013, 8, e79052. [Google Scholar] [CrossRef]
- Madrid-Espinoza, J.; Brunel-Saldias, N.; Guerra, F.P.; Gutiérrez, A.; Del Pozo, A. Genome-wide identification and transcriptional regulation of aquaporin genes in bread wheat (Triticum aestivum L.) under water stress. Genes 2018, 9, 497. [Google Scholar] [CrossRef]
- Bao, F.; Ding, A.; Cheng, T.; Wang, J.; Zhang, Q. Genome-wide analysis of members of the wrky gene family and their cold stress response in Prunus mume. Genes 2019, 10, 911. [Google Scholar] [CrossRef]
- Ke, L.; Lei, W.; Yang, W.; Wang, J.; Gao, J.; Cheng, J.; Sun, Y.; Fan, Z.; Yu, D. Genome-wide identification of cold responsive transcription factors in Brassica napus L. BMC Plant Biol. 2020, 20, 62. [Google Scholar] [CrossRef]
- Dudhate, A.; Shinde, H.; Yu, P.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Comprehensive analysis of NAC transcription factor family uncovers drought and salinity stress response in pearl millet (Pennisetum glaucum). BMC Genom. 2021, 22, 70. [Google Scholar] [CrossRef]
- Musavizadeh, Z.; Najafi-Zarrini, H.; Kazemitabar, S.K.; Hashemi, S.H.; Faraji, S.; Barcaccia, G.; Heidari, P. Genome-wide analysis of potassium channel genes in rice: Expression of the OsAKT and OsKAT genes under salt stress. Genes 2021, 12, 784. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Shinde, H.; Dudhate, A.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Genome-Wide Investigation of SQUAMOSA Promoter Binding Protein-like Transcription Factor Family in Pearl Millet (Pennisetum glaucum (L.) R. Br.). Plant Gene 2021, 27, 100313. [Google Scholar] [CrossRef]
- Li, G.; Jin, L.; Sheng, S. Genome-wide identification of bHLH transcription factor in Medicago sativa in response to cold stress. Genes 2022, 13, 2371. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bishnu, P.K.; Linde, M.; Debener, T.; Alkio, M.; Knoche, M. Expression of putative aquaporin genes in sweet cherry is higher in flesh than skin and most are downregulated during development. Sci. Hortic. 2019, 244, 304–314. [Google Scholar] [CrossRef]
- Afzal, Z.; Howton, T.C.; Sun, Y.; Mukhtar, M.S. The roles of aquaporins in plant stress responses. J. Dev. Biol. 2016, 4, 9. [Google Scholar] [CrossRef]
- Zhu, Y.-X.; Yang, L.; Liu, N.; Yang, J.; Zhou, X.-K.; Xia, Y.-C.; He, Y.; He, Y.-Q.; Gong, H.-J.; Ma, D.-F.; et al. Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biol. 2019, 19, 345. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Ali, Z.; Wang, C.B.; Xu, L.; Yi, J.X.; Xu, Z.L.; Liu, X.Q.; He, X.L.; Huang, Y.H.; Khan, I.A.; et al. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 2013, 8, e56312. [Google Scholar] [CrossRef]
- Zou, Z.; Gong, J.; An, F.; Xie, G.; Wang, J.; Mo, Y.; Yang, L. Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genom. 2015, 16, 1001. [Google Scholar] [CrossRef]
- Ariani, A.; Gepts, P. Genome-wide identification and characterization of aquaporin gene family in common bean (Phaseolus vulgaris L.). Mol. Genet. Genom. 2015, 290, 1771–1785. [Google Scholar] [CrossRef]
- Martins, C.; Pedrosa, A.M.; Du, D.; Gonçalves, L.P.; Yu, Q.; Gmitter, F.G.; Costa, M.G. Genome-wide characterization and expression analysis of Major Intrinsic Proteins during abiotic and biotic stresses in sweet orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0138786. [Google Scholar]
- Hu, W.; Hou, X.; Huang, C.; Yan, Y.; Tie, W.; Ding, Z.; Wei, Y.; Liu, J.; Miao, H.; Lu, Z.; et al. Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 2015, 16, 19728–19751. [Google Scholar] [CrossRef]
- Reddy, P.S.; Rao, T.S.R.B.; Sharma, K.K.; Vadez, V. Genome-wide identification and characterization of the aquaporin gene family in Sorghum bicolor (L.). Plant Gene 2015, 1, 18–28. [Google Scholar] [CrossRef]
- Chaumont, F.; Barrieu, F.; Wojcik, E.; Chrispeels, M.J.; Jung, R. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 2001, 125, 1206–1215. [Google Scholar] [CrossRef]
- Johanson, U.; Karlsson, M.; Johansson, I.; Gustavsson, S.; Sjövall, S.; Fraysse, L.; Weig, A.R.; Kjellbom, P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Phys. 2001, 126, 1358–1369. [Google Scholar] [CrossRef]
- Tao, P.; Zhong, X.; Li, B.; Wang, W.; Yue, Z.; Lei, J. Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. 2014, 289, 1131–1145. [Google Scholar] [CrossRef]
- Li, G.; Santoni, V.; Maurel, C. Plant aquaporins: Roles in plant physiology. Biochim. Biophys. Acta 2014, 1840, 1574–1582. [Google Scholar] [CrossRef]
- Flexas, J.; Ribas-Carbó, M.; Hanson, D.T.; Bota, J.; Otto, B.; Cifre, J.; McDowell, N.; Medrano, H.; Kaldenhoff, R. Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. Plant J. 2006, 48, 427–439. [Google Scholar] [CrossRef]
- Mori, I.C.; Rhee, J.; Shibasaka, M.; Sasano, S.; Kaneko, T.; Horie, T.; Katsuhara, M. CO2 transport by PIP2 aquaporins of barley. Plant Cell Physiol. 2014, 55, 251–257. [Google Scholar] [CrossRef]
- Xu, F.; Wang, K.; Yuan, W.; Xu, W.; Shuang, L.; Kronzucker, H.J.; Chen, G.; Miao, R.; Zhang, M.; Ding, M.; et al. Overexpression of rice aquaporin OsPIP1;2 improves yield by enhancing mesophyll CO2 conductance and phloem sucrose transport. J. Exp. Bot. 2018, 70, 671–681. [Google Scholar] [CrossRef]
- Wang, X.; Gao, F.; Bing, J.; Sun, W.; Feng, X.; Ma, X.; Zhou, Y.; Zhang, G. Overexpression of the jojoba aquaporin gene, ScPIP1, enhances drought and salt tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2019, 20, 153. [Google Scholar] [CrossRef]
- Mosa, K.A.; Kumar, K.; Chhikara, S.; Musante, C.; White, J.C.; Dhankher, O.P. Enhanced boron tolerance in plants mediated by bidirectional transport through plasma membrane intrinsic proteins. Sci. Rep. 2016, 6, 21640. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Mosa, K.A.; Chhikara, S.; Musante, C.; White, J.C.; Dhankher, O.P. Two rice plasma membrane intrinsic proteins, OsPIP2;4 and OsPIP2;7, are involved in transport and providing tolerance to boron toxicity. Planta 2014, 239, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Rhee, J.Y.; Chung, G.C.; Kang, H. Aquaporin as a membrane transporter of hydrogen peroxide in plant response to stresses. Plant Signal. Behav. 2012, 7, 1180–1181. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, M.; Bousser, A.; Sissoeff, I.; Roche, O.; Hoarau, J.; Mahe, A. Cloning and characterization of ZmPIP1-5b, an aquaporin transporting water and urea. Plant Sci. 2003, 165, 21–31. [Google Scholar] [CrossRef]
- Frick, A.; Järvå, M.; Ekvall, M.; Uzdavinys, P.; Nyblom, M.; Törnroth-Horsefield, S. Mercury increases water permeability of a plant aquaporin through a non-cysteine-related mechanism. Biochem. J. 2013, 454, 491–499. [Google Scholar] [CrossRef]
- Wang, H.; Schoebel, S.; Schmitz, F.; Dong, H.; Hedfalk, K. Quantitative analysis of H2O2 transport through purified membrane proteins. MethodsX 2020, 7, 100816. [Google Scholar] [CrossRef]
- Wallace, I.S.; Roberts, D.M. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Phys. 2004, 135, 1059–1068. [Google Scholar] [CrossRef]
- Dean, R.M.; Rivers, R.L.; Zeidel, M.L.; Roberts, D.M. Purification and functional reconstitution of soybean nodulin 26. An aquaporin with water and glycerol transport properties. Biochemistry 1999, 38, 347–353. [Google Scholar] [CrossRef]
- Niemietz, C.M.; Tyerman, S.D. Channel-mediated permeation of ammonia gas through the peribacteroid membrane of soybean nodules. FEBS Lett. 2000, 465, 110–114. [Google Scholar] [CrossRef]
- Wallace, I.S.; Wills, D.M.; Guenther, J.F.; Roberts, D.M. Functional selectivity for glycerol of the nodulin 26 subfamily of plant membrane intrinsic proteins. FEBS Lett. 2002, 523, 109–112. [Google Scholar] [CrossRef]
- Wallace, I.S.; Roberts, D.M. Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 2005, 44, 16826–16834. [Google Scholar] [CrossRef]
- Ma, J.F.; Tamai, K.; Yamaji, N.; Mitani, N.; Konishi, S.; Katsuhara, M.; Ishiguro, M.; Murata, Y.; Yano, M. A silicon transporter in rice. Nature 2006, 440, 688–691. [Google Scholar] [CrossRef]
- Takano, J.; Wada, M.; Ludewig, U.; Schaaf, G.; von Wirén, N.; Fujiwara, T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 2006, 18, 1498–1509. [Google Scholar] [CrossRef]
- Choi, W.G.; Roberts, D.M. Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. J. Biol. Chem 2007, 282, 24209–24218. [Google Scholar] [CrossRef]
- Tanaka, M.; Fujiwara, T. Physiological roles and transport mechanisms of boron: Perspectives from plants. Pflug. Arch. 2008, 456, 671–677. [Google Scholar] [CrossRef]
- Yang, H.; Menz, J.; Häussermann, I.; Benz, M.; Fujiwara, T.; Ludewig, U. High and Low Affinity Urea Root Uptake: Involvement of NIP5;1. Plant Cell Physiol. 2015, 56, 1588–1597. [Google Scholar] [CrossRef]
- Gu, R.; Chen, X.; Zhou, Y.; Yuan, L. Isolation and characterization of three maize aquaporin genes, ZmNIP2;1, ZmNIP2;4 and ZmTIP4;4 involved in urea transport. BMB Rep. 2012, 45, 96–101. [Google Scholar] [CrossRef]
- Soto, G.; Alleva, K.; Mazzella, M.A.; Amodeo, G.; Muschietti, J.P. AtTIP1; 3 and AtTIP5; 1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 2008, 582, 4077–4082. [Google Scholar] [CrossRef]
- Soto, G.; Fox, R.; Ayub, N.; Alleva, K.; Guaimas, F.; Erijman, E.J.; Mazzella, A.; Amodeo, G.; Muschietti, J. TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant J. 2010, 64, 1038–1047. [Google Scholar] [CrossRef]
- Azad, A.K.; Yoshikawa, N.; Ishikawa, T.; Sawa, Y.; Shibata, H. Substitution of a single amino acid residue in the aromatic/arginine selectivity filter alters the transport profiles of tonoplast aquaporin homologs. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1–11. [Google Scholar] [CrossRef]
- Dynowski, M.; Mayer, M.; Moran, O.; Ludewig, U. Molecular determinants of ammonia and urea conductance in plant aquaporin homologs. FEBS Lett. 2008, 582, 2458–2462. [Google Scholar] [CrossRef]
- Ishikawa, F.; Suga, S.; Uemura, T.; Sato, M.H.; Maeshima, M. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 2005, 579, 5814–5820. [Google Scholar] [CrossRef] [PubMed]
- Noronha, H.; Agasse, A.; Martins, A.P.; Berny, M.C.; Gomes, D.; Zarrouk, O.; Gerós, H. The grape aquaporin VvSIP1 transports water across the ER membrane. J. Exp. Bot. 2013, 65, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Ampah-Korsah, H.; Anderberg, H.I.; Engfors, A.; Kirscht, A.; Nordén, K.; Kjellstrom, S.; Johanson, U. The aquaporin splice variant NbXIP1; 1α is permeable to boric acid and is phosphorylated in the N-terminal domain. Front. Plant Sci. 2016, 7, 862. [Google Scholar] [CrossRef]
- Pal, D.; Chakrabarti, P. Non-hydrogen bond interactions involving the methionine sulfur atom. J. Biomol. Struct. Dyn. 2001, 19, 115–128. [Google Scholar] [CrossRef]
- Biswal, H.S.; Gloaguen, E.; Loquais, Y.; Tardivel, B.; Mons, M. Strength of NH··· S hydrogen bonds in methionine residues revealed by gas-phase IR/UV spectroscopy. J. Phys. Chem. 2012, 3, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Yu, J.W.; Park, S.W. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol. Biochem. 2013, 73, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Yang, S.; Wang, Y.; Bendahmane, M.; Fu, X. Genome-wide identification and characterization of aquaporin gene family in Beta vulgaris. Peer J. 2017, 5, e3747. [Google Scholar] [CrossRef]
- Song, S.; Zhang, D.; Ma, F.; Xing, W.; Huang, D.; Wu, B.; Chen, J.; Chen, D.; Xu, B.; Xu, Y. Genome-Wide identification and expression analyses of the aquaporin gene family in passion fruit (Passiflora edulis), revealing petip3-2 to be involved in drought stress. Int. J. Mol. Sci. 2022, 23, 5720. [Google Scholar] [CrossRef]
- Grondin, A.; Mauleon, R.; Vadez, V.; Henry, A. Root aquaporins contribute to whole plant water fluxes under drought stress in rice (Oryza sativa L.). Plant Cell Environ. 2016, 39, 347–365. [Google Scholar] [CrossRef]
- Nguyen, M.X.; Moon, S.; Jung, K.H. Genome-wide expression analysis of rice aquaporin genes and development of a functional gene network mediated by aquaporin expression in roots. Planta 2013, 238, 669–681. [Google Scholar] [CrossRef]
- Ludevid, D.; Höfte, H.; Himelblau, E.; Chrispeels, M.J. The expression pattern of the tonoplast intrinsic protein γ-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol. 1992, 100, 1633–1639. [Google Scholar] [CrossRef]
- Hachez, C.; Heinen, R.B.; Draye, X.; Chaumont, F. The expression pattern of plasma membrane aquaporins in maize leaf highlights their role in hydraulic regulation. Plant Mol. Biol. 2008, 68, 337–353. [Google Scholar] [CrossRef]
- Besse, M.; Knipfer, T.; Miller, A.J.; Verdeil, J.L.; Jahn, T.P.; Fricke, W. Developmental pattern of aquaporin expression in barley (Hordeum vulgare L.). leaves. J. Exp. Bot. 2011, 62, 4127–4142. [Google Scholar] [CrossRef]
- Li, Q.; Tong, T.; Jiang, W.; Cheng, J.; Deng, F.; Wu, X.; Chen, Z.-H.; Ouyang, Y.; Zeng, F. Highly conserved evolution of aquaporin PIPs and TIPs confers their crucial contribution to flowering process in plants. Front. Plant Sci. 2022, 12, 761713. [Google Scholar] [CrossRef]
- Alexandersson, E.; Fraysse, L.; Sjövall-Larsen, S.; Gustavsson, S.; Fellert, M.; Karlsson, M. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol 2005, 59, 469–484. [Google Scholar] [CrossRef]
- Xue, J.; Yang, F.; Gao, J. Isolation of Rh-TIP1;1, an aquaporin gene and its expression in rose flowers in response to ethylene and water deficit. Postharvest. Biol. Technol. 2009, 51, 407–413. [Google Scholar] [CrossRef]
- Tong, Z.; Li, Q.; Yang, Y.; Dai, F.; Gao, J.; Hong, B. Isolation and expression analysis of LoPIP2, a lily (Lilium Oriental Hybrids) aquaporin gene involved in desiccation-induced anther dehiscence. Sci. Hortic. 2013, 164, 316–322. [Google Scholar] [CrossRef]
- Rasheed-Depardieu, C.; Parelle, J.; Tatin-Froux, F.; Parent, C.; Capelli, N. Short-term response to waterlogging in Quercus petraea and Quercus robur: A study of the root hydraulic responses and the transcriptional pattern of aquaporins. Plant Physiol. Biochem. 2015, 97, 323–330. [Google Scholar] [CrossRef]
- North, G.B.; Martre, P.; Nobel, P.S. Aquaporins account for variations in hydraulic conductance for metabolically active root regions of Agave deserti in wet, dry, and rewetted soil. Plant Cell Environ. 2004, 27, 219–228. [Google Scholar] [CrossRef]
- Siemens, J.A.; Zwiazek, J.J. Changes in water flow properties of solution culture-grown trembling aspen (Populus tremuloides) seedlings under different intensities of water-deficit stress. Physiol. Plant 2004, 121, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Mahdieh, M.; Mostajeran, A.; Horie, T.; Katsuhara, M. Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum Plants. Plant Cell Physiol. 2008, 49, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, M.M.; Wiecha, K.; Gajek, K.; Szarejko, I. Drought stress and re-watering affect the abundance of TIP aquaporin transcripts in barley. PLoS ONE 2019, 14, e0226423. [Google Scholar] [CrossRef] [PubMed]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular mycorrhizal fungi in alleviation of salt stress: A review. Ann. Bot. 2009, 104, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Katsuhara, M.; Rhee, J.Y.; Sugimoto, G.; Chung, G.C. Early response in water relations influenced by NaCl reflects tolerance or sensitivity of barley plants to salinity stress via aquaporins. Soil Sci. Plant Nutr. 2011, 57, 50–60. [Google Scholar] [CrossRef]
- Muries, B.; Faize, M.; Carvajal, M.; Martínez-Ballesta, M. Identification and differential induction of the expression of aquaporins by salinity in broccoli plants. Mol Biosyst. 2011, 7, 1322–1335. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Beck, E.H.; Heim, R.; Hansen, J. Plant resistance to cold stress: Mechanisms and environmental signals triggering frost hardening and dehardening. J. Biosci. 2004, 29, 449–459. [Google Scholar] [CrossRef]
- Rahman, A.; Kawamura, Y.; Maeshima, M.; Rahman, A.; Uemura, M. Plasma membrane aquaporin members PIPs act in concert to regulate cold acclimation and freezing tolerance responses in Arabidopsis thaliana. Plant Cell Physiol. 2020, 61, 787–802. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Xing, D.; Gao, C. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J. Exp. Bot. 2009, 7, 2073–2091. [Google Scholar] [CrossRef]
- Grover, A.; Mittal, D.; Negi, M.; Lavania, D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013, 205–206, 38–47. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Obaid, A.Y.; Sabir, J.S.; Atef, A.; Liu, X.; Edris, S.; El-Domyati, F.M.; Jansen, R.K. Analysis of transcriptional response to heat stress in Rhazya stricta. BMC Plant Biol. 2016, 16, 252. [Google Scholar] [CrossRef]


| SF | AQP | AA | NPA I | NPA II | ar/R | WP | PP | TDP | Mw | Ip | FR | Ka/Ks | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PruavNIP1;1 | 281 | NPA | NPA | WVAR | Plas | CM | 6 | 29.77 | 9.30 | FSAYI | 0.6 | 0.432 | |
| PruavNIP2;1 | 291 | NPA | NPA | GSGR | Plas | CM | 6 | 30.91 | 8.64 | LTAYV | 0.63 | 0.287 | |
| PruavNIP4;1 | 267 | NPA | NPA | WVAR | Plas | CM | 6 | 28.18 | 6.41 | LSAYF | 0.17 | 0.722 | |
| NIP | PruavNIP4:2 | 248 | NPA | NPA | WVAR | Plas | CM | 6 | 26.06 | 6.40 | LSAYI | 0.19 | 0.732 |
| PruavNIP5;1 | 298 | NPS | NPI | SIGR | Plas | CM | 6 | 31.26 | 7.64 | FTAYI | 0.11 | 0.517 | |
| PruavNIP5;2 | 280 | NPS | NPV | A-GR | Vacu | CM | 5 | 29.28 | 8.85 | FTAYL | 0.58 | 0.298 | |
| PruavNIP6;1 | 307 | NPS | NPV | TIAR | Plas | CM | 6 | 31.89 | 8.34 | FTAYL | 0.14 | 0.415 | |
| PruavNIP7;1 | 300 | NPA | NPA | AVGR | Plas | CM | 6 | 31.99 | 5.82 | YSAYI | 0.23 | 0.535 | |
| PruavPIP1;1 | 286 | NPA | NPA | FHTR | Plas | CM | 5 | 30.79 | 9.28 | ESAFW | 0.45 | 0.282 | |
| PruavPIP1;2 | 290 | NPA | NPA | FIGR | Plas | CM | 6 | 30.85 | 9.20 | QSAFW | 0.36 | 0.379 | |
| PruavPIP1;3 | 286 | NPA | NPA | FHTR | Plas | CM | 6 | 30.66 | 9.25 | QSAFW | 0.31 | 0.331 | |
| PIP | PruavPIP2;1 | 287 | NPA | NPA | FHTR | Plas | CM | 6 | 30.52 | 8.94 | QSAFW | 0.25 | 0.511 |
| PruavPIP2;2 | 281 | NPA | NPA | FHTR | Plas | CM | 6 | 30.04 | 7.00 | QSAFW | 0.38 | 0.420 | |
| PruavPIP2;3 | 284 | NPA | NPA | FHTR | Plas | CM | 6 | 30.07 | 6.89 | QSAFW | 0.29 | 0.505 | |
| PruavPIP2;4 | 281 | NPA | NPA | FHTR | Plas | CM | 6 | 29.83 | 9.00 | QSAFW | 0.41 | 0.415 | |
| PruavSIP1;1 | 240 | NPS | NPA | AVPN | Vacu | CM.VC | 5 | 25.18 | 9.82 | MAAYW | 0.35 | 0.896 | |
| SIP | PruavSIP1;2 | 244 | NPT | NPA | TVPN | Chlo | CM.VC | 6 | 25.88 | 9.52 | MAAYW | 0.22 | 0.755 |
| PruavSIP2;1 | 236 | NPL | NPA | SHGS | Plas | CM | 4 | 25.80 | 9.52 | FVAYW | 0.53 | 0.538 | |
| PruavTIP1;1 | 252 | NPA | NPA | HIAV | Plas | VC | 6 | 25.94 | 5.54 | TSAYW | 0.18 | 0.769 | |
| PruavTIP1;2 | 252 | NPA | NPA | HIAV | Vacu | VC | 6 | 26.08 | 4.78 | TSAYW | 0.25 | 0.884 | |
| PruavTIP1;3 | 252 | NPA | NPA | HIAV | Plas | VC | 6 | 26.15 | 6.12 | TSAYW | 0.41 | 0.735 | |
| TIP | PruavTIP2;1 | 248 | NPA | NPA | HIGR | Plas | VC | 7 | 25.40 | 6.26 | TSAYW | 0.55 | 0.941 |
| PruavTIP2;2 | 248 | NPA | NPA | HIGR | Plas | VC | 6 | 25.17 | 4.86 | TSAYW | 0.44 | 0.982 | |
| PruavTIP3;1 | 256 | NPA | NPA | HIGR | Plas | VC | 6 | 27.26 | 7.11 | TAAYW | 0.58 | 0.530 | |
| PruavTIP4;1 | 249 | NPA | NPA | HIAR | Vacu | VC | 7 | 26.24 | 5.54 | TSAYW | 0.54 | 0.821 | |
| PruavTIP5;1 | 255 | NPA | NPA | NVGC | Plas | CM.VC | 6 | 25.84 | 6.81 | VAAYW | 0.06 | 0.842 | |
| XIP | PruavXIP1;1 | 304 | SLV | SPA | VIVR | Plas | CM | 6 | 32.17 | 5.97 | MCAFW | 0.43 | 0.725 |
| PruavXIP2;1 | 314 | NPV | NPA | ITVR | Plas | CM | 7 | 33.85 | 7.74 | VCAFW | 0.55 | 0.665 |
| Substrate | Aquaporin | Specificity-Determining Positions | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SDP1 | SDP2 | SDP3 | SDP4 | SDP5 | SDP6 | SDP7 | SDP8 | SDP9 | ||
| Ammonia Transporter | F/T | K/L/N/V | F/T | V/L/T | A | D/S | A/H/L | E/P/S | A/R/T | |
| PruavTIP2;1 | T | L | I | L | A | T | H | P | V | |
| Boric Acid Transporter | T/V | I/V | H/I | P | E | I/L | I/L/T | A/T | A/G/K/P | |
| PruavPIP1:1 | T | I | H | P | E | L | L | T | P | |
| PruavPIP1:2 | T | I | H | P | E | L | L | T | P | |
| PruavPIP1:3 | T | I | H | P | E | L | L | T | P | |
| PruavPIP2;1 | T | I | H | P | E | I | L | T | P | |
| PruavPIP2;2 | T | I | H | P | E | I | L | T | P | |
| PruavPIP2;3 | T | I | H | P | E | I | L | T | P | |
| PruavPIP2;4 | T | I | H | P | E | I | L | T | P | |
| PruavNIP1;1 | V | I | H | P | E | L | M | A | P | |
| PruavNIP2;1 | V | I | H | P | E | I | I | A | P | |
| PruavNIP4;1 | V | I | H | P | E | I | F | A | P | |
| PruavNIP4;2 | V | I | H | P | E | L | F | A | P | |
| PruavNIP5;1 | V | I | H | P | E | V | L | A | P | |
| PruavNIP5;2 | V | I | H | P | E | L | L | A | P | |
| PruavNIP6;1 | V | I | H | P | E | L | L | A | E | |
| PruavNIP7;1 | I | I | H | P | E | L | L | T | P | |
| PruavXIP2;1 | T | I | H | P | E | I | T | T | V | |
| CO2 Transporter | I/L/V | I | C | A | I/V | D | W | D | W | |
| PruavPIP1;1 | V | I | T | A | I | D | W | D | W | |
| PruavPIP1;3 | V | I | T | A | I | D | W | D | W | |
| PruavPIP2;2 | V | I | S | A | V | D | W | D | W | |
| H2O2 Transporter | A/S | A/G | L/V | A/F/L/T/V | I/L/V | H/I/L/Q | F/Y | A/V | P | |
| PruavNIP2;1 | S | A | L | L | V | I | Y | V | P | |
| PruavNIP4;1 | S | A | L | L | V | L | Y | A | P | |
| PruavNIP5;1 | S | A | L | V | I | L | Y | V | P | |
| PruavPIP1;1 | A | G | V | F | I | H | F | V | P | |
| PruavPIP1;2 | A | G | V | F | I | H | F | V | P | |
| PruavPIP1;3 | A | G | V | F | I | H | F | V | P | |
| PruavPIP2;1 | A | G | V | F | I | Q | F | V | P | |
| PruavPIP2;2 | A | G | V | F | I | Q | F | L | P | |
| PruavPIP2;3 | A | G | V | F | I | Q | F | V | P | |
| PruavPIP2;4 | A | G | V | I | I | Q | F | V | P | |
| PruavTIP1;1 | S | A | L | A | I | H | Y | A | P | |
| PruavTIP1;3 | A | A | L | V | I | H | Y | V | P | |
| PruavTIP2;1 | A | A | L | V | I | N | Y | V | P | |
| PruavTIP2;2 | S | A | L | V | I | N | Y | V | P | |
| PruavTIP3;1 | A | A | L | V | I | H | Y | V | P | |
| PruavTIP5;1 | S | A | L | T | I | Q | Y | V | P | |
| PruavXIP2;1 | A | G | L | V | V | H | F | V | P | |
| Silicic acid Transporter | C/S | F/Y | A/E/L | H/R/Y | G | K/N/T | R | E/S/T | A/K/P/T | |
| PruavNIP2;1 | S | F | V | H | G | N | R | S | N | |
| Urea Transporter | H | P | F/I/L/T | A/C/F/L | L/M | A/G/P | G/S | G/S | N | |
| PruavNIP1;1 | H | P | I | A | L | P | G | S | N | |
| PruavNIP2;1 | H | P | L | A | M | P | G | S | N | |
| PruavNIP5;1 | H | P | I | A | L | P | G | S | N | |
| PruavNIP5;2 | H | P | I | A | L | P | G | S | N | |
| PruavPIP1;1 | H | P | F | F | L | P | G | G | N | |
| PruavPIP1;2 | H | P | F | F | L | P | G | G | N | |
| PruavPIP1;3 | H | P | F | F | L | P | G | G | N | |
| PruavPIP2;1 | H | P | F | F | L | P | G | G | N | |
| PruavPIP2;2 | H | P | F | F | L | P | G | G | N | |
| PruavPIP2;3 | H | P | F | F | L | P | G | G | N | |
| PruavPIP2;4 | H | P | F | F | L | P | G | G | N | |
| PruavTIP1;1 | H | P | F | F | L | A | G | S | N | |
| PruavTIP1;2 | H | P | F | F | L | A | G | S | N | |
| PruavTIP1;3 | H | P | F | A | L | P | G | S | N | |
| PruavTIP2;1 | H | P | F | A | L | P | G | S | N | |
| PruavTIP2;2 | H | P | L | A | L | P | G | S | N | |
| PruavTIP3;1 | H | P | F | L | L | P | G | S | N | |
| PruavTIP4;1 | H | P | L | L | L | A | G | S | N | |
| PruavTIP5;1 | H | P | F | A | L | P | G | S | N | |
| PruavXIP1;1 | H | L | F | A | V | G | G | G | N | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvatierra, A.; Mateluna, P.; Toro, G.; Solís, S.; Pimentel, P. Genome-Wide Identification and Gene Expression Analysis of Sweet Cherry Aquaporins (Prunus avium L.) under Abiotic Stresses. Genes 2023, 14, 940. https://doi.org/10.3390/genes14040940
Salvatierra A, Mateluna P, Toro G, Solís S, Pimentel P. Genome-Wide Identification and Gene Expression Analysis of Sweet Cherry Aquaporins (Prunus avium L.) under Abiotic Stresses. Genes. 2023; 14(4):940. https://doi.org/10.3390/genes14040940
Chicago/Turabian StyleSalvatierra, Ariel, Patricio Mateluna, Guillermo Toro, Simón Solís, and Paula Pimentel. 2023. "Genome-Wide Identification and Gene Expression Analysis of Sweet Cherry Aquaporins (Prunus avium L.) under Abiotic Stresses" Genes 14, no. 4: 940. https://doi.org/10.3390/genes14040940
APA StyleSalvatierra, A., Mateluna, P., Toro, G., Solís, S., & Pimentel, P. (2023). Genome-Wide Identification and Gene Expression Analysis of Sweet Cherry Aquaporins (Prunus avium L.) under Abiotic Stresses. Genes, 14(4), 940. https://doi.org/10.3390/genes14040940