Unraveling the Dysbiosis of Vaginal Microbiome to Understand Cervical Cancer Disease Etiology—An Explainable AI Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Bioinformatic Processing and Statistical Analysis
2.3. SHAP Interpretation of Vaginal Microbiome Associated with Cervical Cancer
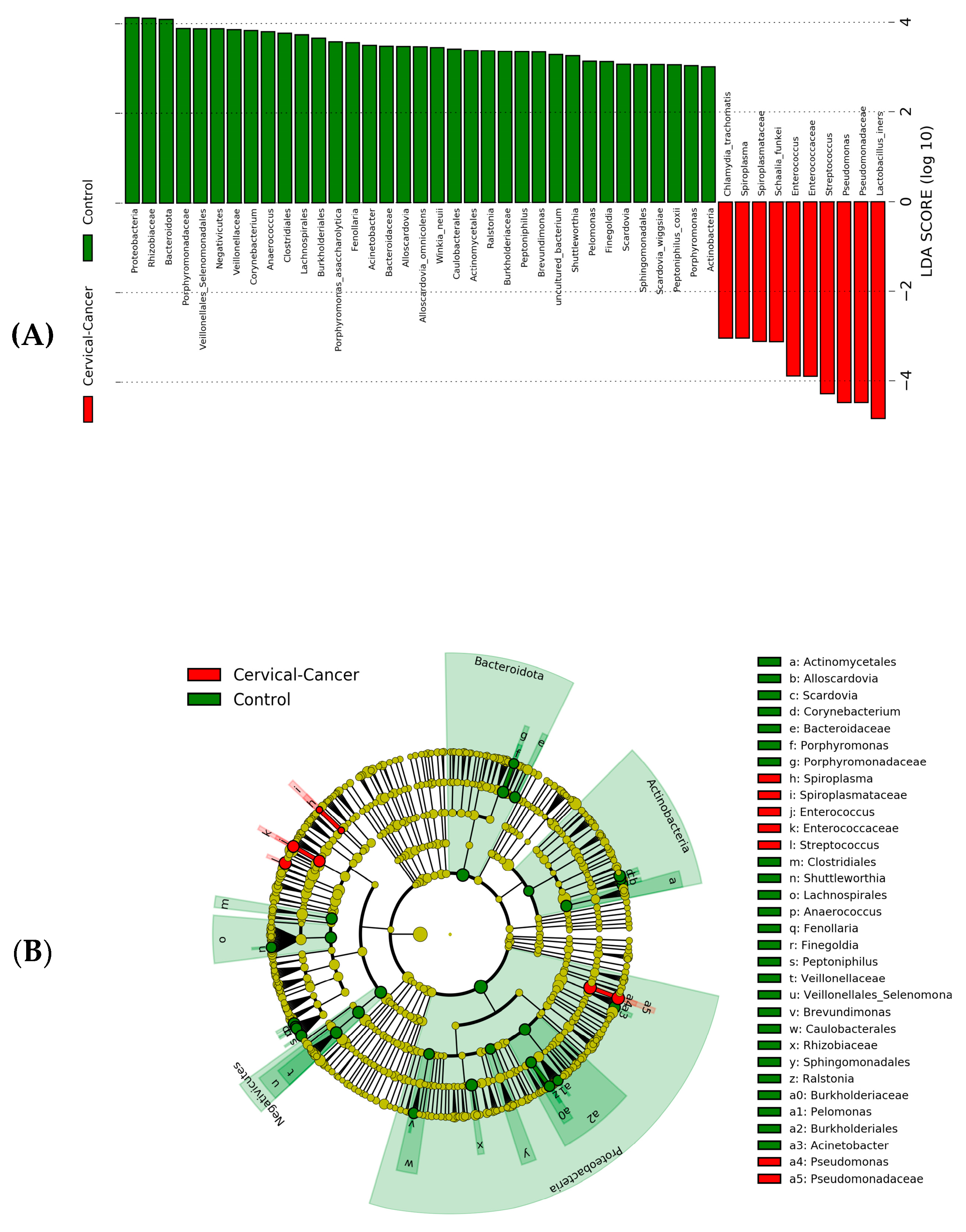
3. Results
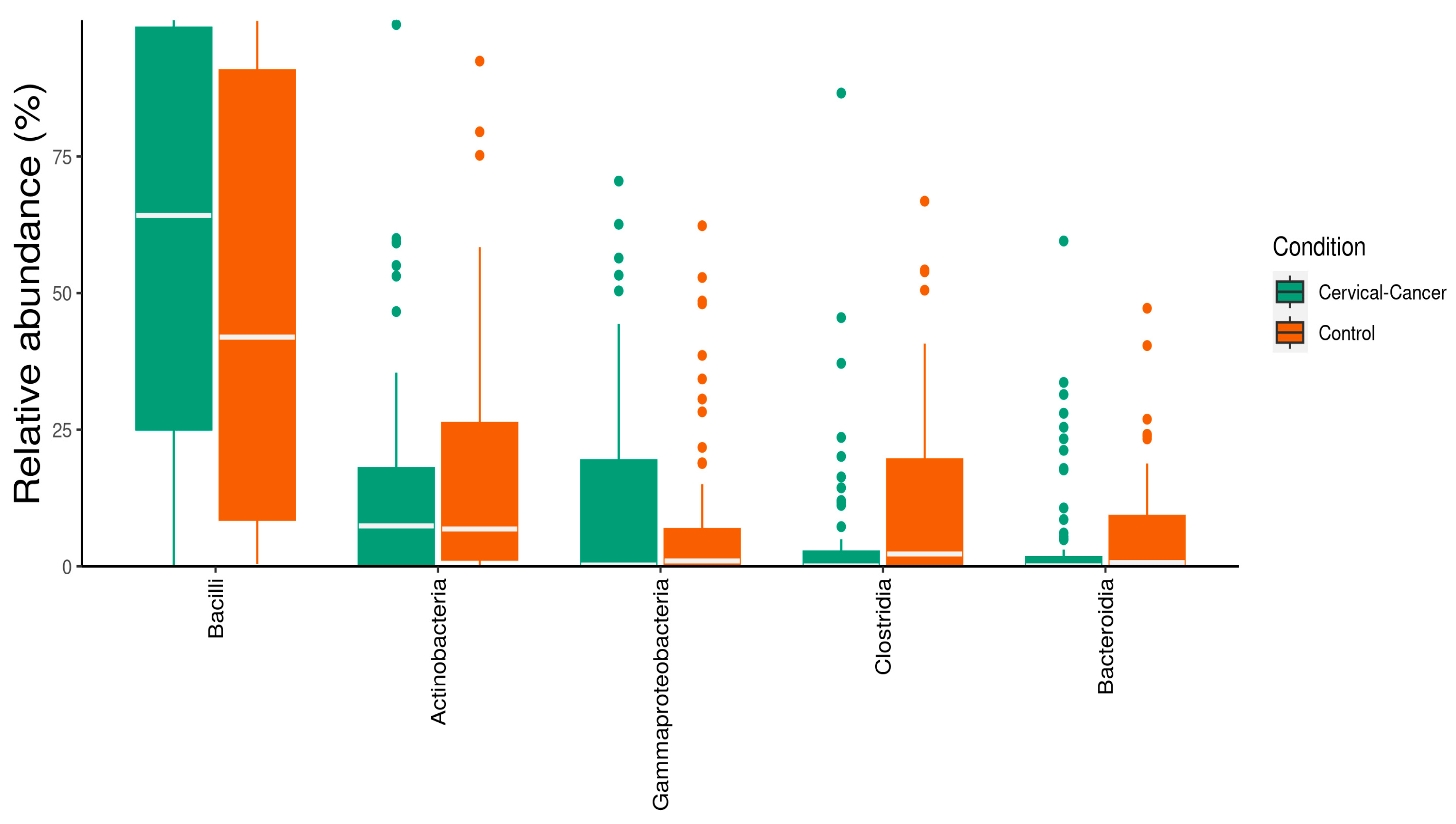
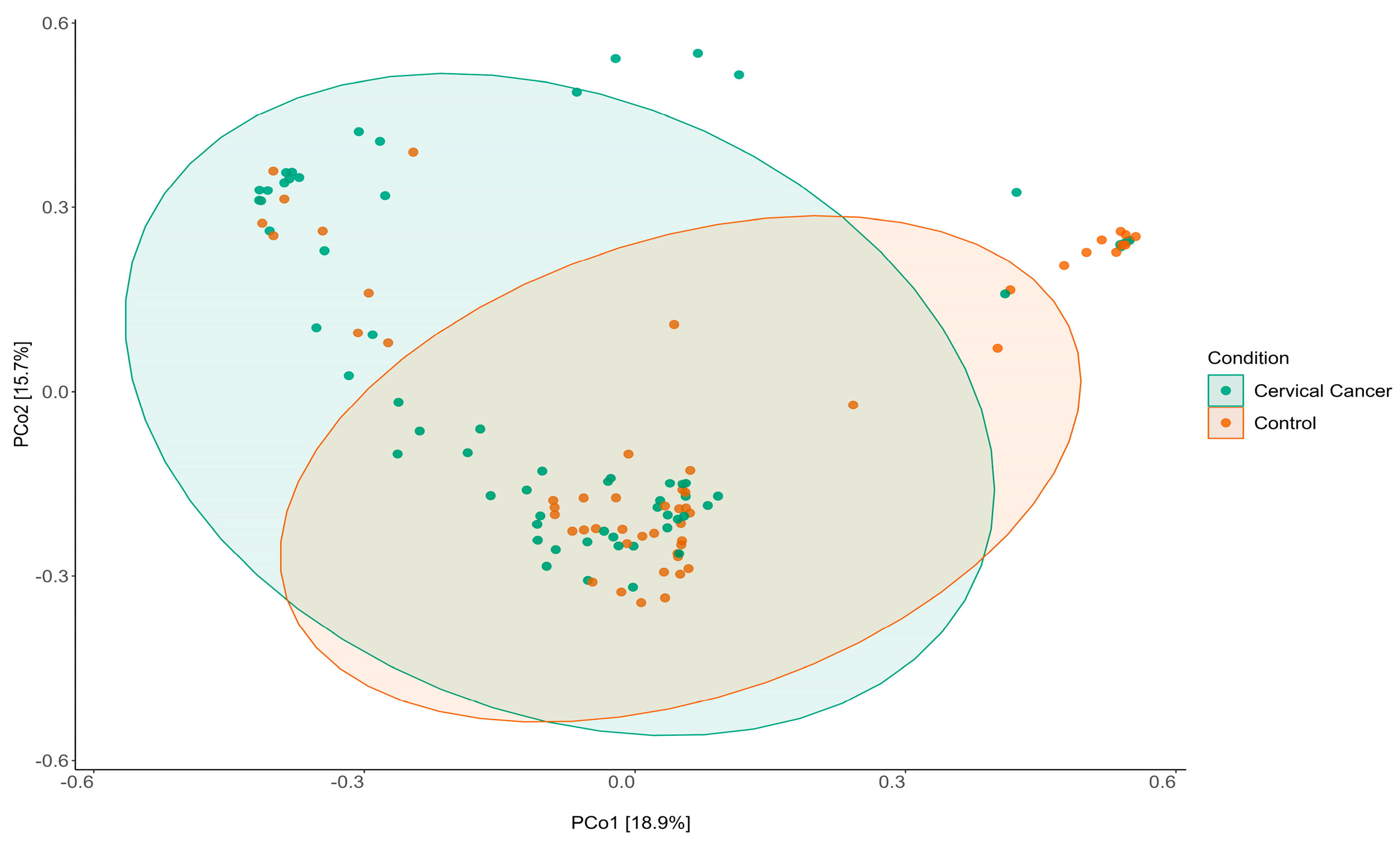
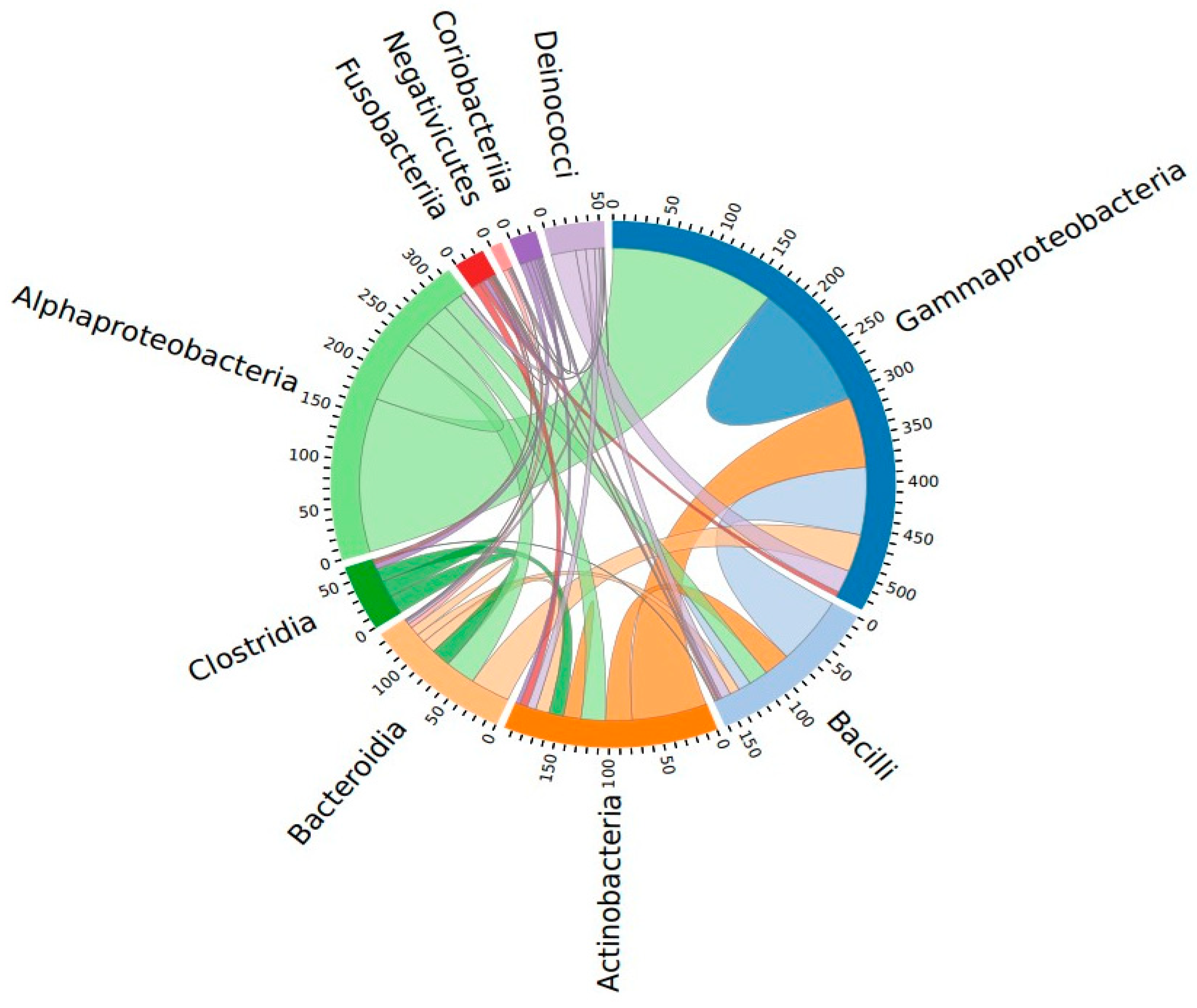
3.1. Characterization of Vaginal Microbiome
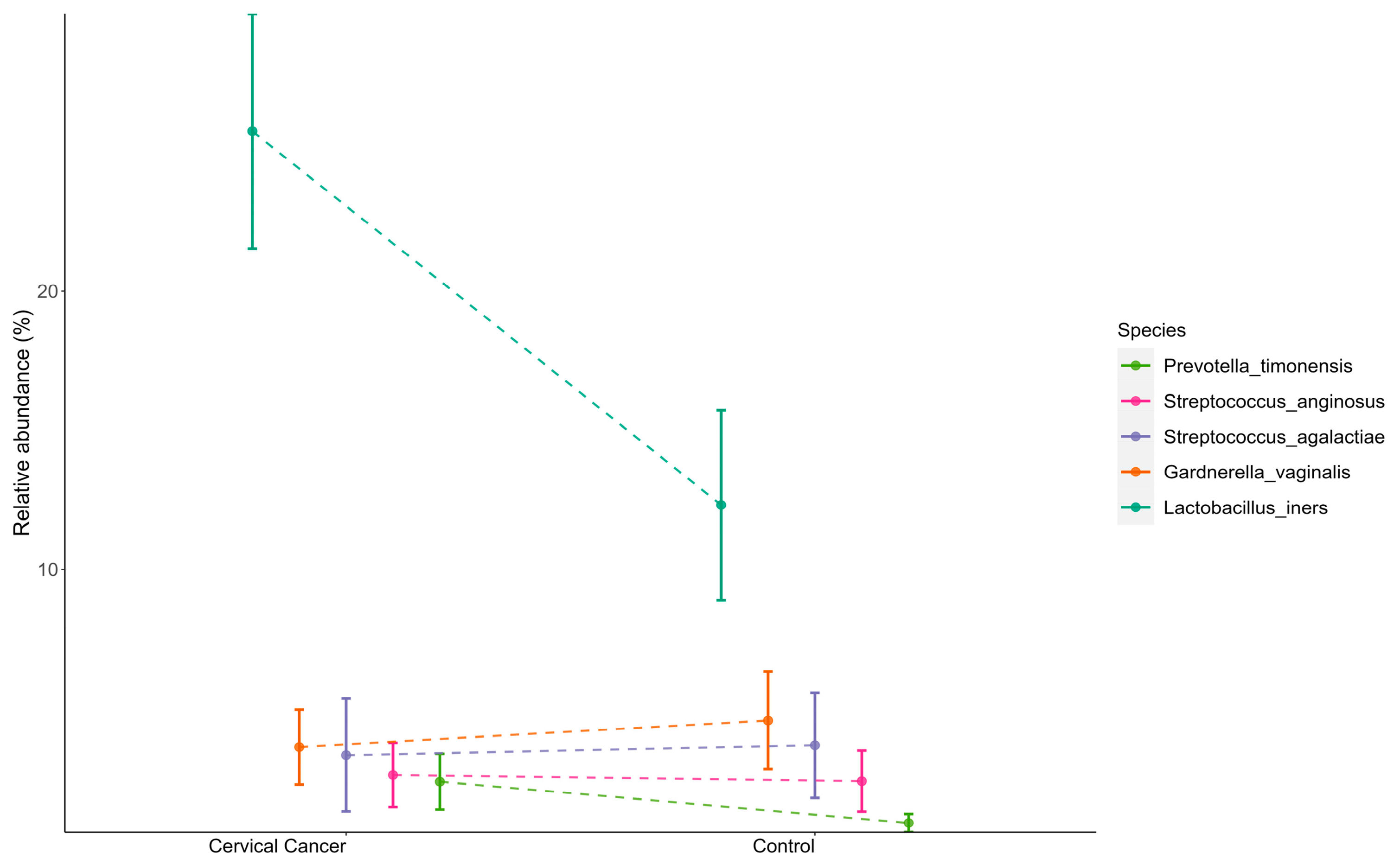
3.2. Dysbiosis of Vaginal Microbiome Associated with Cervical Cancer
3.3. Explaining the Model Predictions through SHAP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Foreman, K.J.; Marquez, N.; Dolgert, A.; Fukutaki, K.; Fullman, N.; McGaughey, M.; Pletcher, M.A.; Smith, A.E.; Tang, K.; Yuan, C.-W.; et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: Reference and alternative scenarios for 2016–40 for 195 countries and territories. Lancet 2018, 392, 2052–2090. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Pimple, S.; Mishra, G. Cancer cervix: Epidemiology and disease burden. CytoJournal 2022, 19, 21. [Google Scholar] [CrossRef]
- William, W.; Ware, A.; Basaza-Ejiri, A.H.; Obungoloch, J. A review of image analysis and machine learning techniques for automated cervical cancer screening from pap-smear images. Comput. Methods Programs Biomed. 2018, 164, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Devi, B.C.R.; Tang, T.S.; Corbex, M. Reducing by half the percentage of late-stage presentation for breast and cervix cancer over 4 years: A pilot study of clinical downstaging in Sarawak, Malaysia. Ann. Oncol. 2007, 18, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Hicks, M.L.; Yap, O.W.S.; Matthews, R.; Parham, G. Disparities in cervical cancer screening, treatment and outcomes. Ethn. Dis. 2006, 16 (Suppl. S3), S3-63–S3-66. [Google Scholar]
- Plummer, M.; Herrero, R.; Franceschi, S.; Meijer, C.J.L.M.; Snijders, P.; Bosch, F.X.; de Sanjosé, S.; Muñoz, N. Smoking and cervical cancer: Pooled analysis of the IARC multi-centric case–control study. Cancer Causes Control 2003, 14, 805–814. [Google Scholar] [CrossRef]
- Roura, E.; Castellsagué, X.; Pawlita, M.; Travier, N.; Waterboer, T.; Margall, N.; Bosch, F.X.; de Sanjosé, S.; Dillner, J.; Gram, I.T.; et al. Smoking as a major risk factor for cervical cancer and pre-cancer: Results from the EPIC cohort. Int. J. Cancer 2014, 135, 453–466. [Google Scholar] [CrossRef]
- Adebamowo, S.N.; Dareng, E.O.; Famooto, A.O.; Offiong, R.; Olaniyan, O.; Obende, K.; Adebayo, A.; Ologun, S.; Alabi, B.; Achara, P.; et al. Cohort Profile: African Collaborative Center for Microbiome and Genomics Research’s (ACCME’s) Human Papillomavirus (HPV) and Cervical Cancer Study. Int. J. Epidemiol. 2017, 46, 1745–1745j. [Google Scholar] [CrossRef]
- Seo, S.-S.; Oh, H.Y.; Lee, J.-K.; Kong, J.-S.; Lee, D.O.; Kim, M.K. Combined effect of diet and cervical microbiome on the risk of cervical intraepithelial neoplasia. Clin. Nutr. 2016, 35, 1434–1441. [Google Scholar] [CrossRef]
- Amabebe, E.; Anumba, D.O.C. The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 2018, 5, 181. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Marchesi, J.R.; Lee, Y.S.; Bennett, P.R.; Kyrgiou, M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: What do we know and where are we going next? Microbiome 2016, 4, 58. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Fettweis, J.M.; Brooks, J.P.; Jefferson, K.K.; Buck, G.A. The changing landscape of the vaginal microbiome. Clin. Lab. Med. 2014, 34, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Audirac-Chalifour, A.; Torres-Poveda, K.; Bahena-Román, M.; Téllez-Sosa, J.; Martínez-Barnetche, J.; Cortina-Ceballos, B.; López-Estrada, G.; Delgado-Romero, K.; Burguete-García, A.I.; Cantú, D.; et al. Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PLoS ONE 2016, 11, e0153274. [Google Scholar] [CrossRef]
- Chase, D.; Goulder, A.; Zenhausern, F.; Monk, B.; Herbst-Kralovetz, M. The vaginal and gastrointestinal microbiomes in gynecologic cancers: A review of applications in etiology, symptoms and treatment. Gynecol. Oncol. 2015, 138, 190–200. [Google Scholar] [CrossRef]
- Łaniewski, P.; Barnes, D.; Goulder, A.; Cui, H.; Roe, D.J.; Chase, D.M.; Herbst-Kralovetz, M.M. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci. Rep. 2018, 8, 7593. [Google Scholar] [CrossRef]
- Łaniewski, P.; Ilhan, Z.E.; Herbst-Kralovetz, M.M. The microbiome and gynaecological cancer development, prevention and therapy. Nat. Rev. Urol. 2020, 17, 232–250. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Lee, Y.S.; Smith, A.; Marchesi, J.R.; Lehne, B.; Bhatia, R.; Lyons, D.; Paraskevaidis, E.; Li, J.V.; et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015, 5, 16865. [Google Scholar] [CrossRef]
- Cheng, L.; Norenhag, J.; Hu, Y.O.O.; Brusselaers, N.; Fransson, E.; Ährlund-Richter, A.; Guðnadóttir, U.; Angelidou, P.; Zha, Y.; Hamsten, M.; et al. Vaginal microbiota and human papillomavirus infection among young Swedish women. Npj Biofilm. Microbiomes 2020, 6, 39. [Google Scholar] [CrossRef]
- Klein, C.; Kahesa, C.; Mwaiselage, J.; West, J.T.; Wood, C.; Angeletti, P.C. How the Cervical Microbiota Contributes to Cervical Cancer Risk in Sub-Saharan Africa. Front. Cell. Infect. Microbiol. 2020, 10, 23. [Google Scholar] [CrossRef]
- Brusselaers, N.; Shrestha, S.; Wijgert, J.v.d.; Verstraelen, H. Vaginal dysbiosis and the risk of human papillomavirus and cervical cancer: Systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2019, 221, 9–18.e8. [Google Scholar] [CrossRef] [PubMed]
- Ravilla, R.; Coleman, H.; Chan, L.; Chow, C.-E.; Fuhrman, B.; Greenfield, W.; Robeson, M.S.; Iverson, K.; Spencer, H.J.; Nakagawa, M. Cervical microbiome role in outcomes of therapeutic HPV vaccination for cervical intraepithelial neoplasia. J. Clin. Oncol. 2018, 36 (Suppl. S15), 3099. [Google Scholar] [CrossRef]
- Tango, C.N.; Seo, S.-S.; Kwon, M.; Lee, D.-O.; Chang, H.K.; Kim, M.K. Taxonomic and Functional Differences in Cervical Microbiome Associated with Cervical Cancer Development. Sci. Rep. 2020, 10, 9720. [Google Scholar] [CrossRef]
- Arokiyaraj, S.; Seo, S.S.; Kwon, M.; Lee, J.K.; Kim, M.K. Association of cervical microbial community with persistence, clearance, and negativity of Human Papillomavirus in Korean women: A longitudinal study. Sci. Rep. 2018, 8, 15479. [Google Scholar] [CrossRef]
- Khan, I.; Nam, M.; Kwon, M.; Seo, S.; Jung, S.; Han, J.S.; Hwang, G.-S.; Kim, M.K. LC/MS-Based Polar Metabolite Profiling Identified Unique Biomarker Signatures for Cervical Cancer and Cervical Intraepithelial Neoplasia Using Global and Targeted Metabolomics. Cancers 2019, 11, 511. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Seo, S.-S.; Kim, M.K.; Lee, D.O.; Lim, M.C. Compositional and Functional Differences between Microbiota and Cervical Carcinogenesis as Identified by Shotgun Metagenomic Sequencing. Cancers 2019, 11, 309. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Yang, X.; He, L.; Yan, S.; Chen, X.; Que, G. The impact of caries status on supragingival plaque and salivary microbiome in children with mixed dentition: A cross-sectional survey. BMC Oral Health 2021, 21, 319. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed]
- Cameron, E.S.; Schmidt, P.J.; Tremblay, B.J.-M.; Emelko, M.B.; Müller, K.M. Enhancing diversity analysis by repeatedly rarefying next generation sequencing data describing microbial communities. Sci. Rep. 2021, 11, 22302. [Google Scholar] [CrossRef] [PubMed]
- Thukral, A.K. A review on measurement of Alpha diversity in biology. Agric. Res. J. 2017, 54, 1. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Baniecki, H.; Kretowicz, W.; Piatyszek, P.; Wisniewski, J.; Biecek, P. Dalex: Responsible machine learning with interactive explainability and fairness in Python. J. Mach. Learn. Res. 2021, 22, 9759–9765. [Google Scholar]
- Fidel, G.; Bitton, R.; Shabtai, A. When explainability meets adversarial learning: Detecting adversarial examples using shap signatures. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–8. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Yang, X.; Da, M.; Zhang, W.; Qi, Q.; Zhang, C.; Han, S. Role of Lactobacillus in cervical cancer. Cancer Manag. Res. 2018, 10, 1219. [Google Scholar] [CrossRef] [PubMed]
- Kyrgiou, M.; Moscicki, A.B. Vaginal microbiome and cervical cancer. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Colbert, L.E.; Karpinets, T.V.; El Alam, M.B.; Lynn, E.J.; Sammouri, J.; Lo, D.; Elnaggar, J.H.; Wang, R.; Harris, T.A.; Yoshida-Court, K.; et al. Cancer-associated Lactobacillus iners are genetically distinct and associated with chemoradiation resistance in cervical cancer. medRxiv 2022. [Google Scholar] [CrossRef]
- Chambers, L.M.; Bussies, P.; Vargas, R.; Esakov, E.; Tewari, S.; Reizes, O.; Michener, C. The microbiome and gynecologic cancer: Current evidence and future opportunities. Curr. Oncol. Rep. 2021, 23, 92. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Shrivastava, S.K.; Joy, L. Cervicovaginal microbiota and HPV-induced cervical cancer. In Immunopathology, Diagnosis and Treatment of HPV Induced Malignancies; Academic Press: Cambridge, MA, USA, 2022; pp. 81–97. [Google Scholar]
- Raffone, A.; Travaglino, A.; Angelino, A.; Esposito, R.; Orlandi, G.; Toscano, P.; Mollo, A.; Insabato, L.; Sansone, M.; Zullo, F. Gardnerella vaginalis and Trichomonas vaginalis infections as risk factors for persistence and progression of low-grade precancerous cervical lesions in HIV-1 positive women. Pathol.-Res. Pract. 2021, 219, 153349. [Google Scholar] [CrossRef]
- Liu, H.; Liang, H.; Li, D.; Wang, M.; Li, Y. Association of Cervical Dysbacteriosis, HPV Oncogene Expression, and Cervical Lesion Progression. Microbiol. Spectr. 2022, 10, e00151-22. [Google Scholar] [CrossRef]
- Wei, W.; Xie, L.Z.; Xia, Q.; Fu, Y.; Liu, F.Y.; Ding, D.N.; Han, F.J. The role of vaginal microecology in the cervical cancer. J. Obstet. Gynaecol. Res. 2022, 48, 2237–2254. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, B.; Lin, Y.; Lin, Y.; Zuo, X. Dysbiosis of Cervical and Vaginal Microbiota Associated with Cervical Intraepithelial Neoplasia. Front. Cell. Infect. Microbiol. 2022, 12, 767693. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, R.; Huang, J.; Qin, X.; Hu, D.; Guo, E.; Liu, C.; Lu, F.; You, L.; Sun, C.; et al. The diversity of vaginal microbiota predicts neoadjuvant chemotherapy responsiveness in locally advanced cervical cancer. Microb. Ecol. 2022, 84, 302–313. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sekaran, K.; Varghese, R.P.; Gopikrishnan, M.; Alsamman, A.M.; El Allali, A.; Zayed, H.; Doss C, G.P. Unraveling the Dysbiosis of Vaginal Microbiome to Understand Cervical Cancer Disease Etiology—An Explainable AI Approach. Genes 2023, 14, 936. https://doi.org/10.3390/genes14040936
Sekaran K, Varghese RP, Gopikrishnan M, Alsamman AM, El Allali A, Zayed H, Doss C GP. Unraveling the Dysbiosis of Vaginal Microbiome to Understand Cervical Cancer Disease Etiology—An Explainable AI Approach. Genes. 2023; 14(4):936. https://doi.org/10.3390/genes14040936
Chicago/Turabian StyleSekaran, Karthik, Rinku Polachirakkal Varghese, Mohanraj Gopikrishnan, Alsamman M. Alsamman, Achraf El Allali, Hatem Zayed, and George Priya Doss C. 2023. "Unraveling the Dysbiosis of Vaginal Microbiome to Understand Cervical Cancer Disease Etiology—An Explainable AI Approach" Genes 14, no. 4: 936. https://doi.org/10.3390/genes14040936
APA StyleSekaran, K., Varghese, R. P., Gopikrishnan, M., Alsamman, A. M., El Allali, A., Zayed, H., & Doss C, G. P. (2023). Unraveling the Dysbiosis of Vaginal Microbiome to Understand Cervical Cancer Disease Etiology—An Explainable AI Approach. Genes, 14(4), 936. https://doi.org/10.3390/genes14040936






