microRNA-660 Enhances Cisplatin Sensitivity via Decreasing SATB2 Expression in Lung Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Clinical Tissue Samples
2.3. Cell Culture
2.4. Plasmids, Mimics/Inhibitor, and Transfection
2.5. RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Assays
2.6. Cisplatin IC50 Detection
2.7. Cell Proliferation
2.8. Colony Formation Assay
2.9. Flow Cytometry
2.10. Luciferase Reporter Assay
2.11. Western Blot
2.12. Statistical Analysis
3. Results
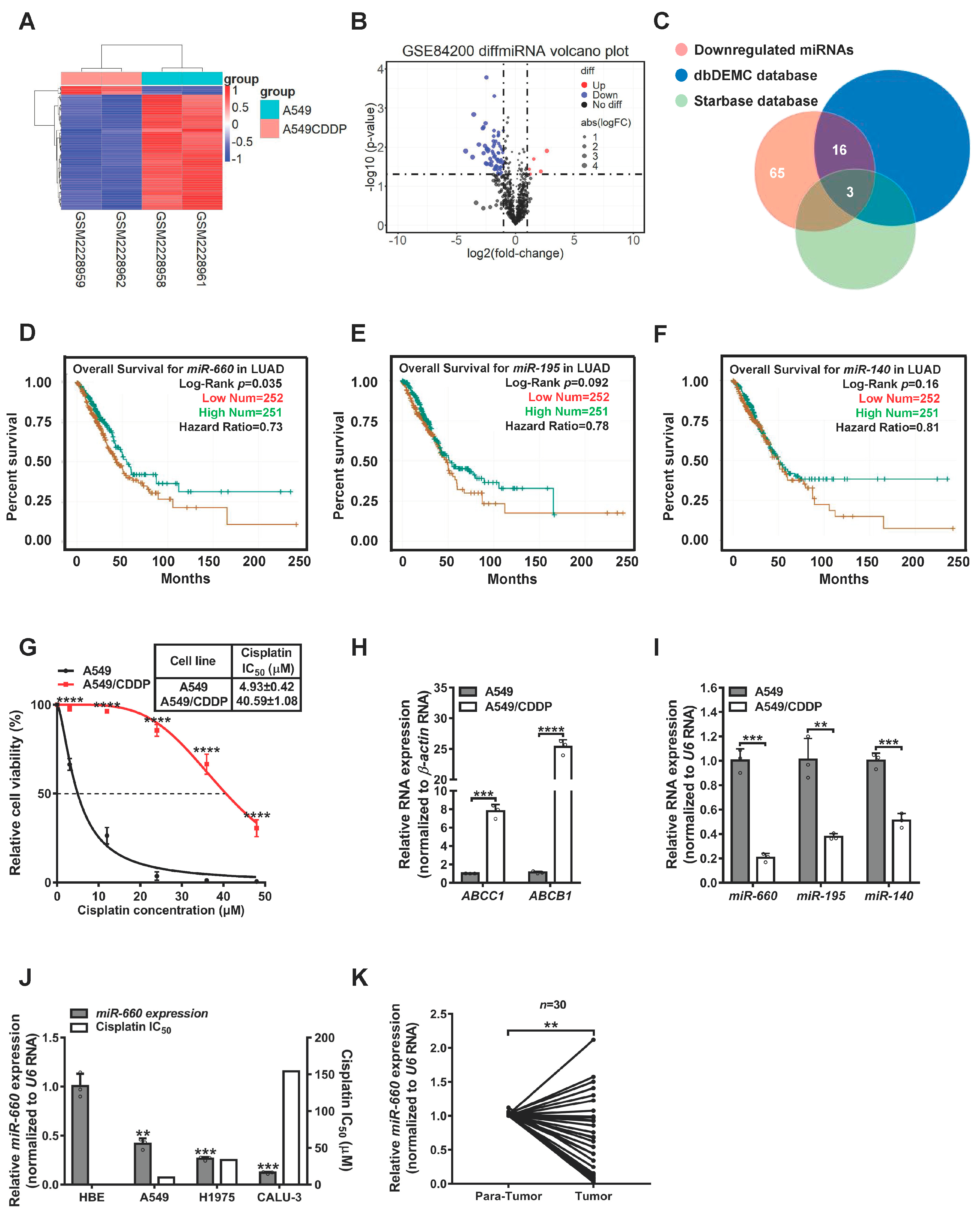
3.1. miR-660 Expression Is Associated with Cisplatin Sensitivity in LUAD Cells
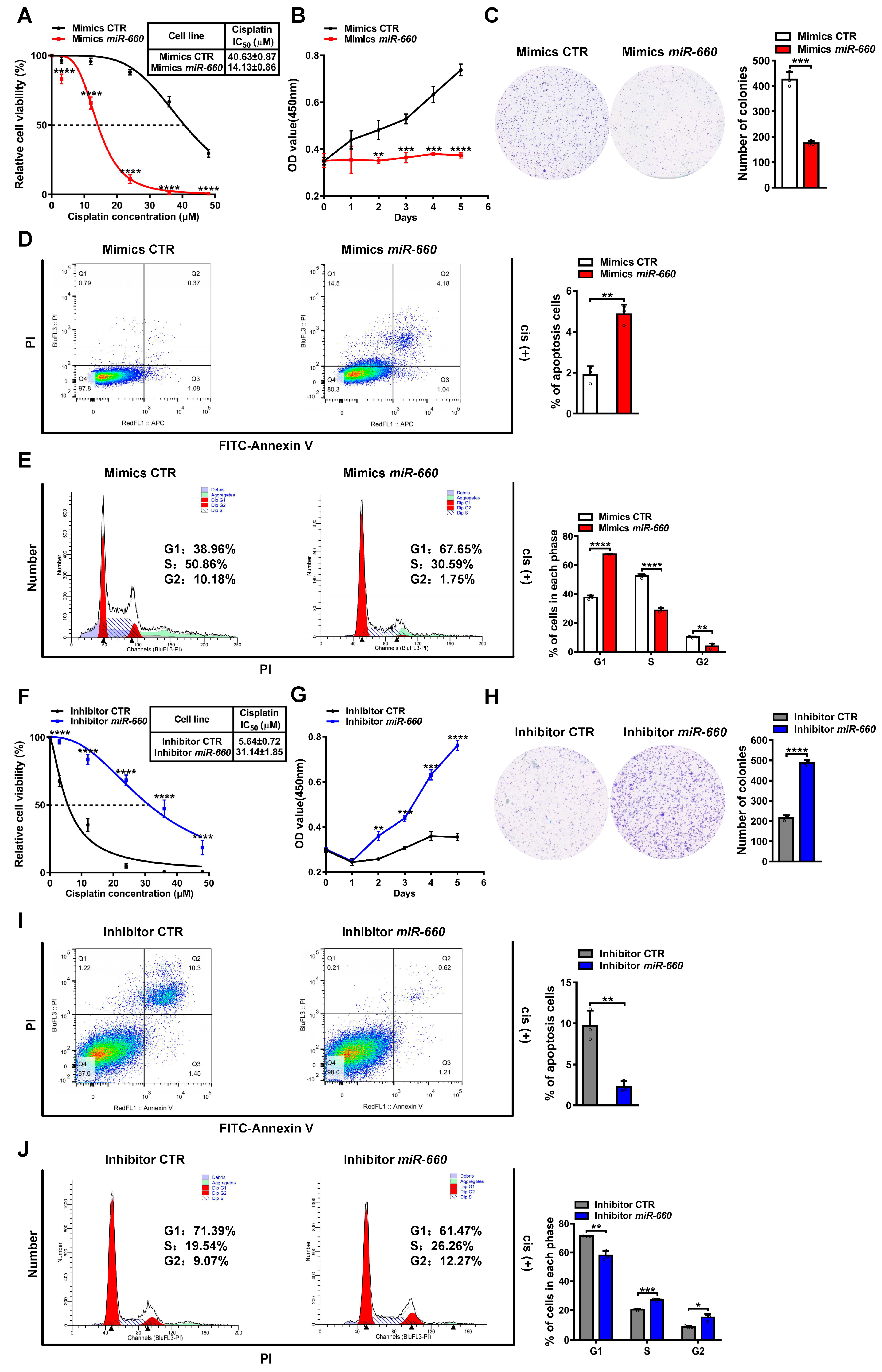
3.2. miR-660 Enhances Cisplatin Sensitivity in LUAD Cells
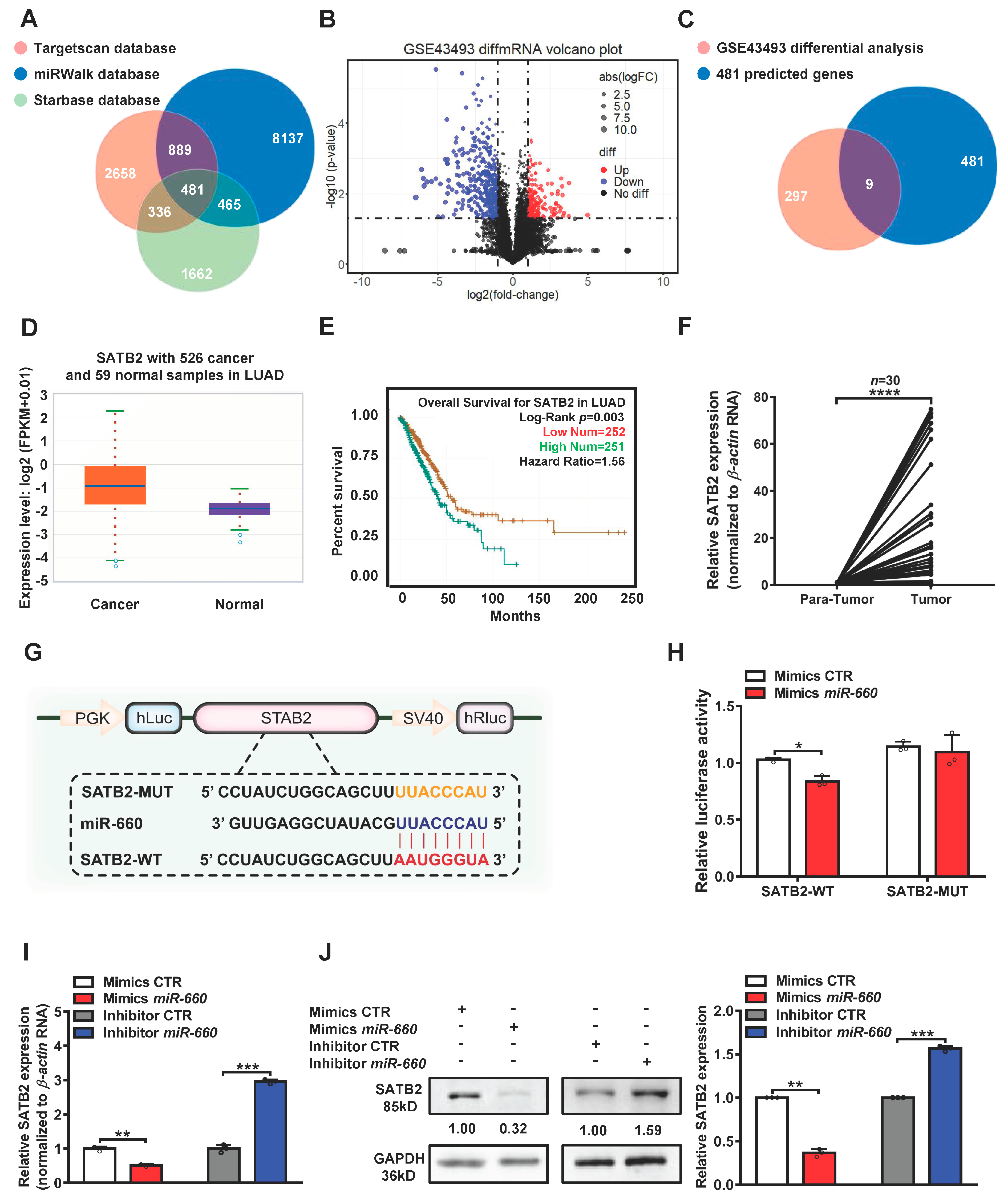
3.3. SATB2 Is a Direct Target of miR-660
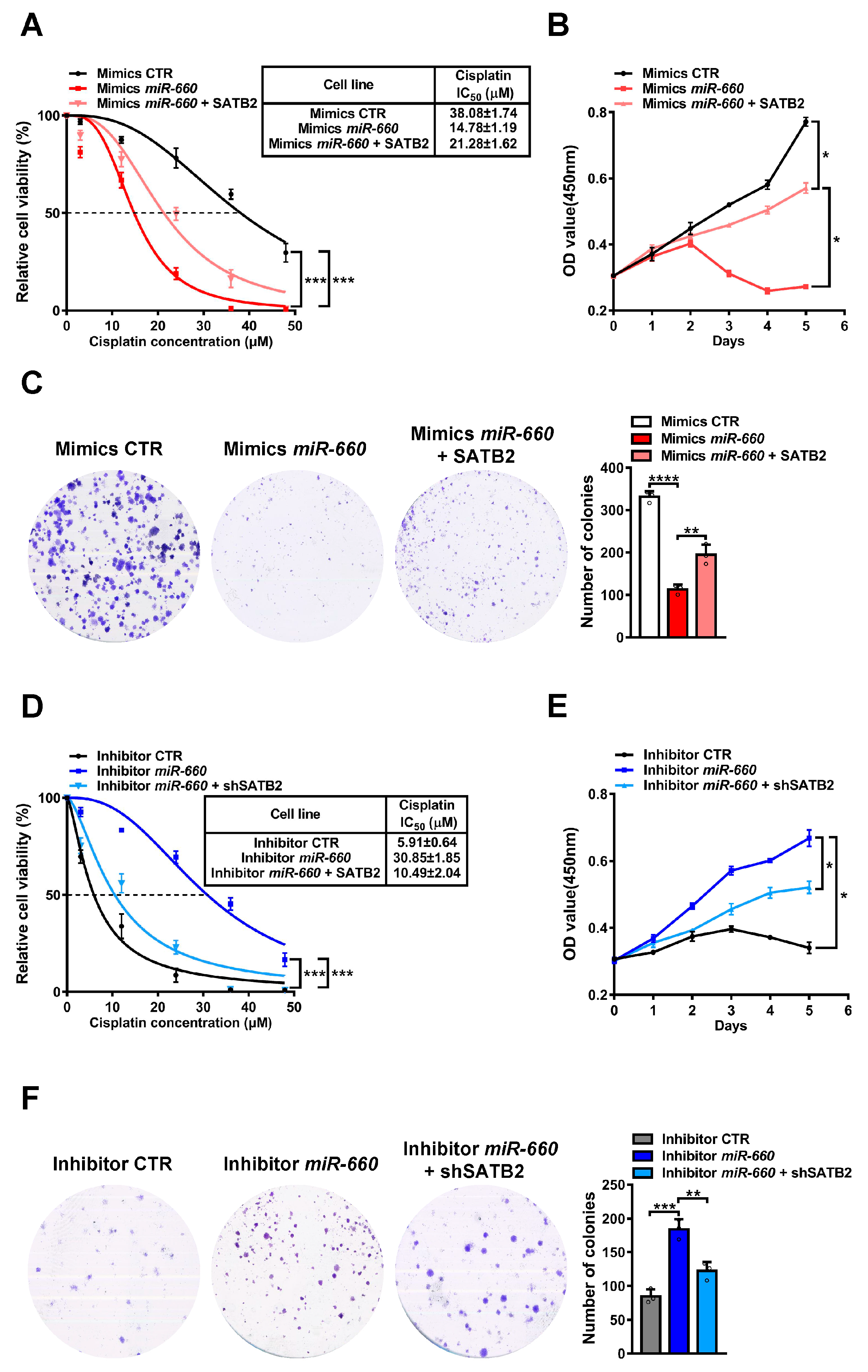
3.4. miR-660 Regulates Cisplatin Sensitivity in LUAD Cells through SATB2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Saab, S.; Zalzale, H.; Rahal, Z.; Khalifeh, Y.; Sinjab, A.; Kadara, H. Insights into Lung Cancer Immune-Based Biology, Prevention, and Treatment. Front. Immunol. 2020, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Pekarek, L.; Navarro, F.; Fraile-Martínez, O.; García-Montero, C.; Álvarez-Mon, M.; Diez-Pedrero, R.; Boyano-Adánez, M.D.C.; Guijarro, L.G.; Barrena-Blázquez, S.; et al. Updated Views in Targeted Therapy in the Patient with Non-Small Cell Lung Cancer. J. Pers. Med. 2023, 13, 167. [Google Scholar] [CrossRef]
- Zulfiqar, B.; Farooq, A.; Kanwal, S.; Asghar, K. Immunotherapy and targeted therapy for lung cancer: Current status and future perspectives. Front. Pharmacol. 2022, 13, 1035171. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tian, L.; Khan, M.N.; Zhang, L.; Chen, Q.; Zhao, Y.; Yan, Q.; Fu, L.; Liu, J. Ginsenoside Rg3 sensitizes hypoxic lung cancer cells to cisplatin via blocking of NF-κB mediated epithelial–mesenchymal transition and stemness. Cancer Lett. 2018, 415, 73–85. [Google Scholar] [CrossRef]
- Hussain, S.; Singh, A.; Nazir, S.U.; Tulsyan, S.; Khan, A.; Kumar, R.; Bashir, N.; Tanwar, P.; Mehrotra, R. Cancer drug resistance: A fleet to conquer. J. Cell. Biochem. 2019, 120, 14213–14225. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ma, J.; Zhou, W.; Zhou, X.; Cao, B.; Zhang, H.; Zhao, Q.; Fan, D.; Hong, L. Molecular mechanisms and clinical implications of miRNAs in drug resistance of esophageal cancer. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef]
- Lei, L.; Huang, Y.; Gong, W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol. Rep. 2013, 30, 2897–2902. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, X.; Chen, L.; Zhao, Y.; Huang, Z.; Zhou, H.; Shi, M. MiR-181a Promotes Apoptosis and Reduces Cisplatin Resistance by Inhibiting Osteopontin in Cervical Cancer Cells. Cancer Biother. Radiopharm. 2019, 34, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Vera, O.; Jimenez, J.; Pernia, O.; Rodriguez-Antolin, C.; Rodriguez, C.; Cabo, F.S.; Soto, J.; Rosas, R.; Lopez-Magallon, S.; Rodriguez, I.E.; et al. DNA Methylation of miR-7 is a Mechanism Involved in Platinum Response through MAFG Overexpression in Cancer Cells. Theranostics 2017, 7, 4118–4134. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, H.; Hou, S.; Hu, B.; Liu, J.; Wang, J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS ONE 2013, 8, e65309. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Xu, F.; Wang, Y.; Ling, Y.; Zhou, C.; Wang, H.; Teschendorff, A.E.; Zhao, Y.; Zhao, H.; He, Y.; Zhang, G.; et al. dbDEMC 3.0: Functional Exploration of Differentially Expressed miRNAs in Cancers of Human and Model Organisms. Genom. Proteom. Bioinform. 2022, 20, 446–454. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012, 41, D955–D961. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, R.; Fang, L.; Ge, X.; Chen, L.; Zhou, M.; Zhou, Y.; Xiong, W.; Hu, Y.; Tang, X.; et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics 2019, 9, 2460–2474. [Google Scholar] [CrossRef]
- Ge, X.; Li, G.-Y.; Jiang, L.; Jia, L.; Zhang, Z.; Li, X.; Wang, R.; Zhou, M.; Zhou, Y.; Zeng, Z.; et al. Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis. Oncogene 2019, 38, 3061–3076. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Kowal, J.; Broude, E.; Roninson, I.; Locher, K.P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 2019, 363, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Nashtahosseini, Z.; Aghamaali, M.R.; Sadeghi, F.; Heydari, N.; Parsian, H. Circulating status of microRNAs 660-5p and 210-3p in breast cancer patients. J. Gene Med. 2021, 23, e3320. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Li, C.; He, L.; Tian, M.; Li, X. miR-660-5p promotes breast cancer progression through down-regulating TET2 and activating PI3K/AKT/mTOR signaling. Braz. J. Med. Biol. Res. 2020, 53, e9740. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Ye, Y.F.; Ruan, L.W.; Bao, L.; Wu, M.W.; Zhou, Y. Inhibition of miR-660-5p expression suppresses tumor development and metastasis in human breast cancer. Genet. Mol. Res. 2017, 16, gmr160194. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Wang, F.; Ni, Q.; Li, M. miR-660-5p promotes the progression of hepatocellular carcinoma by interaction with YWHAH via PI3K/Akt signaling pathway. Biochem. Biophys. Res. Commun. 2020, 531, 480–489. [Google Scholar] [CrossRef]
- Moro, M.; Di Paolo, D.; Milione, M.; Centonze, G.; Bornaghi, V.; Borzi, C.; Gandellini, P.; Perri, P.; Pastorino, U.; Ponzoni, M.; et al. Coated cationic lipid-nanoparticles entrapping miR-660 inhibit tumor growth in patient-derived xenografts lung cancer models. J. Control. Release 2019, 308, 44–56. [Google Scholar] [CrossRef]
- Ai, C.; Ma, G.; Deng, Y.; Zheng, Q.; Gen, Y.; Li, W.; Li, Y.; Zu, L.; Zhou, Q. Nm23-H1 inhibits lung cancer bone-specific metastasis by upregulating miR-660-5p targeted SMARCA5. Thorac. Cancer 2020, 11, 640–650. [Google Scholar] [CrossRef]
- Hu, C.; Xia, R.; Zhang, X.; Li, T.; Ye, Y.; Li, G.; He, R.; Li, Z.; Lin, Q.; Zheng, S.; et al. circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol. Cancer 2022, 21, 24. [Google Scholar] [CrossRef]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef]
- Bi, G.; Liang, J.; Zhao, M.; Zhang, H.; Jin, X.; Lu, T.; Zheng, Y.; Bian, Y.; Chen, Z.; Huang, Y.; et al. miR-6077 promotes cisplatin/pemetrexed resistance in lung adenocarcinoma via CDKN1A/cell cycle arrest and KEAP1/ferroptosis pathways. Mol. Ther. Nucleic Acids 2022, 28, 366–386. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Chen, W.; Chen, K.; Zhu, S.; Lin, F.; Qi, Y.; Zhang, Y.; Han, S.; Rao, T.; Ruan, Y.; et al. TFAP2C Knockdown Sensitizes Bladder Cancer Cells to Cisplatin Treatment via Regulation of EGFR and NF-κB. Cancers 2022, 14, 4809. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhu, Y.; Xu, Q.; Chen, S.; Huang, Y.; Zhao, G.; Ni, X.; Liu, B.; Zhao, W.; Yin, X. YTHDF2 promotes intrahepatic cholangiocarcinoma progression and desensitises cisplatin treatment by increasing CDKN1B mRNA degradation. Clin. Transl. Med. 2022, 12, e848. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Ma, Y.; Shankar, S.; Srivastava, R.K. Role of SATB2 in human pancreatic cancer: Implications in transformation and a promising biomarker. Oncotarget 2016, 7, 57783–57797. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Shankar, S.; Srivastava, R.K. SATB2/β-catenin/TCF-LEF pathway induces cellular transformation by generating cancer stem cells in colorectal cancer. Sci. Rep. 2017, 7, 10939. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, C.F.; Sun, L.B.; Li, Y.C. microRNA-4270-5p inhibits cancer cell proliferation and metastasis in hepatocellular carcinoma by targeting SATB2. Hum. Cell 2020, 33, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Li, Z.; Wang, Y.; Ju, X.; Huang, R. miR-34a Inhibits Cell Proliferation by Targeting SATB2 in Hepatocellular Carcinoma. BioMed Res. Int. 2018, 2018, 2863902. [Google Scholar] [CrossRef]
- Magnusson, K.; de Wit, M.; Brennan, D.J.; Johnson, L.B.; McGee, S.F.; Lundberg, E.; Naicker, K.; Klinger, R.; Kampf, C.; Asplund, A.; et al. SATB2 in Combination with Cytokeratin 20 Identifies Over 95% of all Colorectal Carcinomas. Am. J. Surg. Pathol. 2011, 35, 937–948. [Google Scholar] [CrossRef]
- Fukuhara, M.; Agnarsdóttir, M.; Edqvist, P.H.; Coter, A.; Ponten, F. SATB2 is expressed in Merkel cell carcinoma. Arch. Dermatol. Res. 2016, 308, 449–454. [Google Scholar] [CrossRef]
- Hoskoppal, D.; Epstein, J.I.; Gown, A.M.; Egloff, S.A.A.; Gordetsky, J.B.; Shi, C.J.; A Giannico, G. SATB2 protein expression by immunohistochemistry is a sensitive and specific marker of appendiceal and rectosigmoid well differentiated neuroendocrine tumours. Histopathology 2020, 76, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Bellizzi, A.M. SATB2 in neuroendocrine neoplasms: Strong expression is restricted to well-differentiated tumours of lower gastrointestinal tract origin and is most frequent in Merkel cell carcinoma among poorly differentiated carcinomas. Histopathology 2020, 76, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Lau, J.; Cheng, L.S.; Grant, R.I.; Robinson, F.; Ketela, T.; Reis, P.P.; Roche, O.; Kamel-Reid, S.; Moffat, J.; et al. SATB2 augments ΔNp63α in head and neck squamous cell carcinoma. EMBO Rep. 2010, 11, 777–783. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhou, L.; Chen, B.; Li, X.; Zou, Q.; Xu, W.; Fang, L.; Wu, A.; Li, Z.; Chen, Y. microRNA-660 Enhances Cisplatin Sensitivity via Decreasing SATB2 Expression in Lung Adenocarcinoma. Genes 2023, 14, 911. https://doi.org/10.3390/genes14040911
Wang Z, Zhou L, Chen B, Li X, Zou Q, Xu W, Fang L, Wu A, Li Z, Chen Y. microRNA-660 Enhances Cisplatin Sensitivity via Decreasing SATB2 Expression in Lung Adenocarcinoma. Genes. 2023; 14(4):911. https://doi.org/10.3390/genes14040911
Chicago/Turabian StyleWang, Ziyao, Lingxuan Zhou, Bisong Chen, Xu Li, Qiuyi Zou, Wei Xu, Li Fang, Anbang Wu, Zheng Li, and Yuejun Chen. 2023. "microRNA-660 Enhances Cisplatin Sensitivity via Decreasing SATB2 Expression in Lung Adenocarcinoma" Genes 14, no. 4: 911. https://doi.org/10.3390/genes14040911
APA StyleWang, Z., Zhou, L., Chen, B., Li, X., Zou, Q., Xu, W., Fang, L., Wu, A., Li, Z., & Chen, Y. (2023). microRNA-660 Enhances Cisplatin Sensitivity via Decreasing SATB2 Expression in Lung Adenocarcinoma. Genes, 14(4), 911. https://doi.org/10.3390/genes14040911





