A Complex Intrachromosomal Rearrangement Disrupting IRF6 in a Family with Popliteal Pterygium and Van der Woude Syndromes
Abstract
1. Introduction
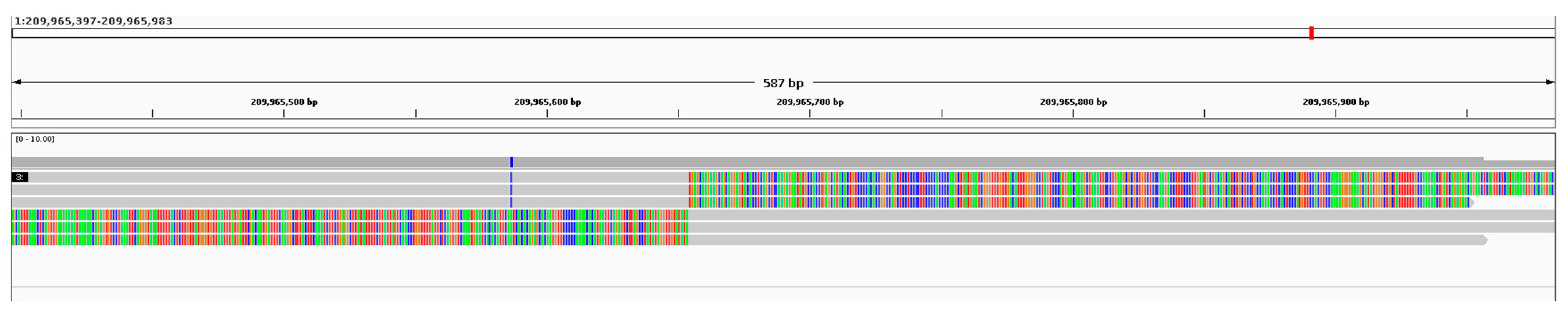
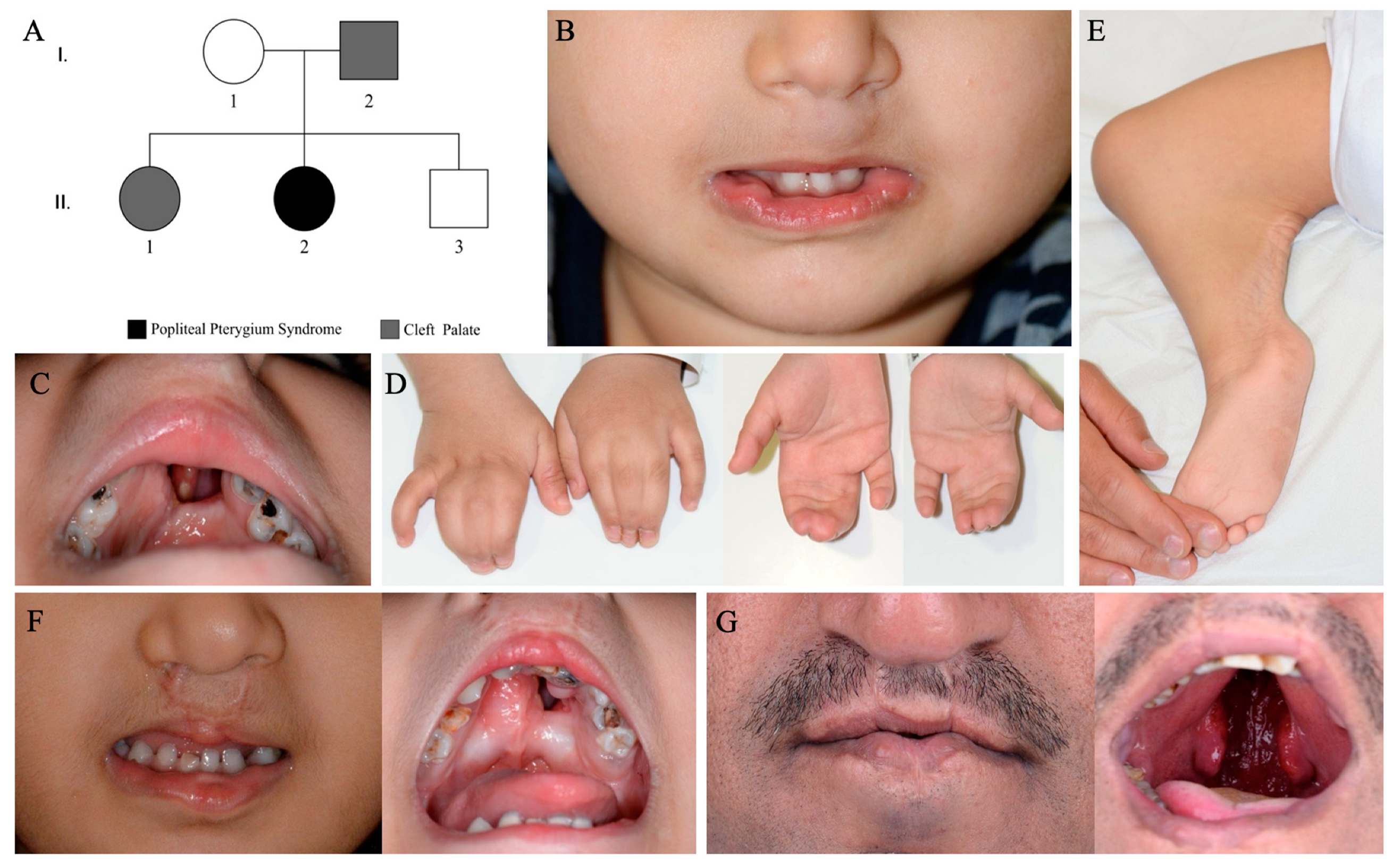
2. Detailed Case Description
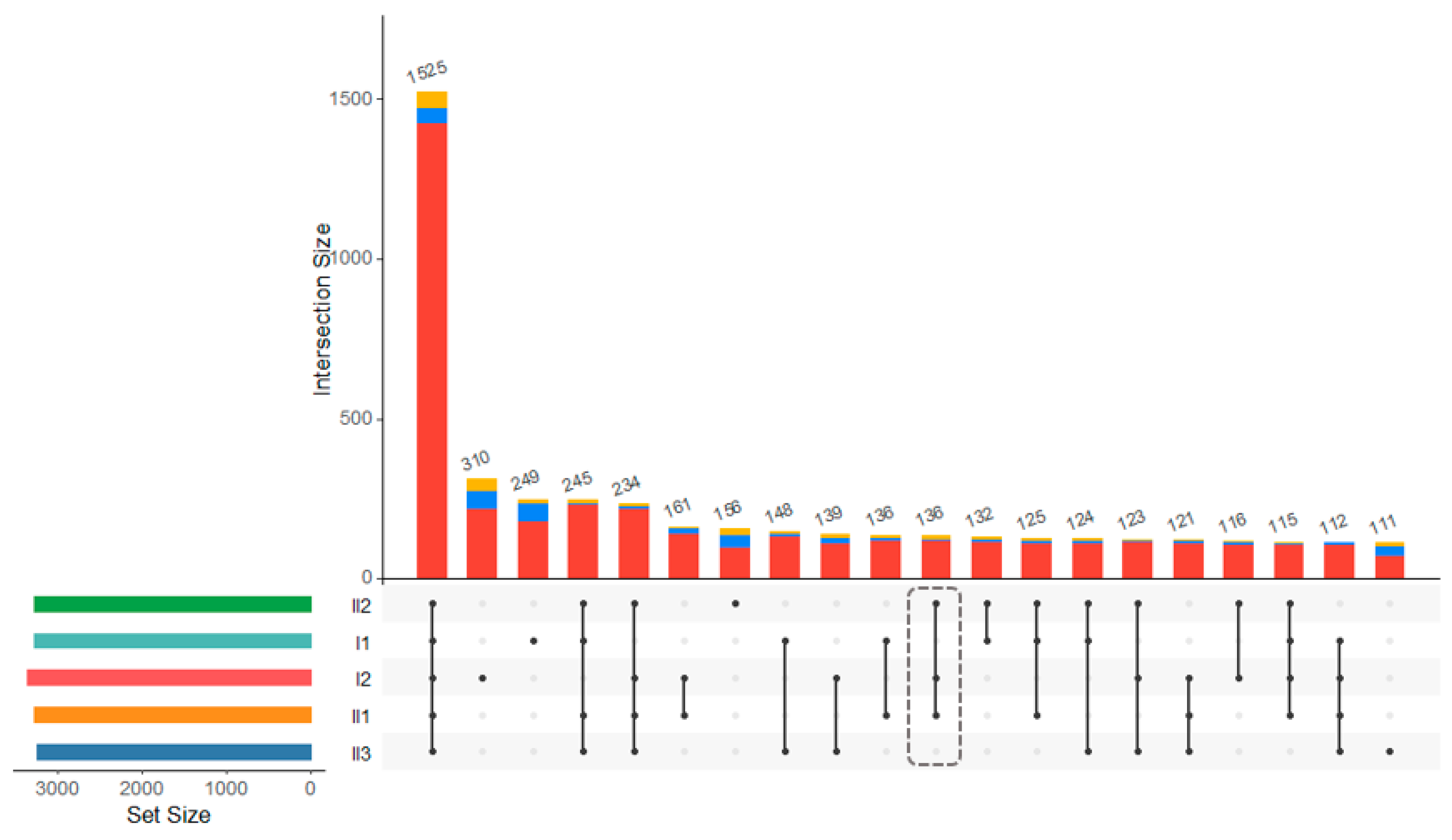
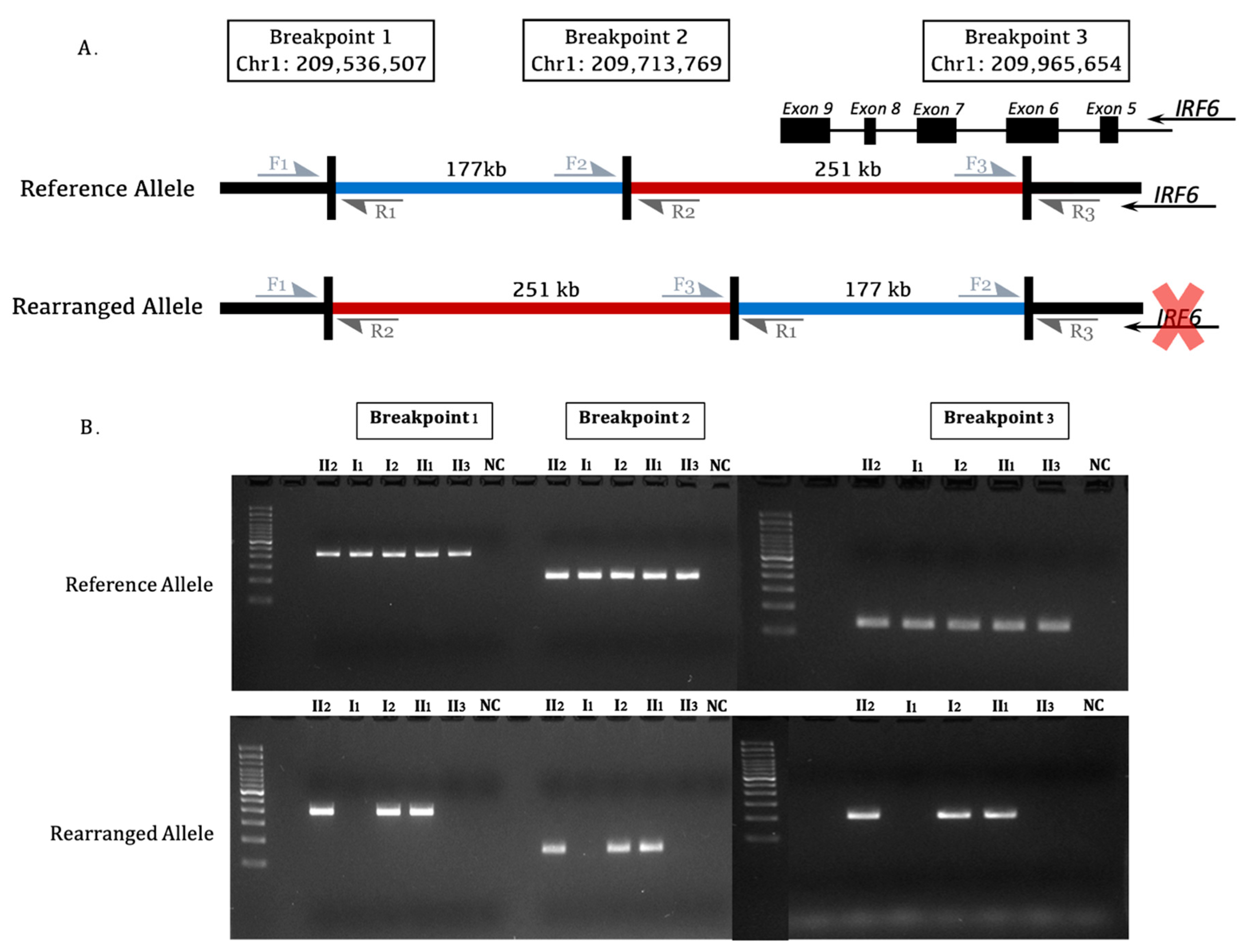
3. Results
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Whole-Genome Sequencing
Appendix A.2. Primer Design

| Primer Set | Forward/Reverse | Sequence 5′ > 3′ | Product Size |
|---|---|---|---|
| P1 | Forward | TCCCAGGGAAGCAGCATTTT | 437 |
| Reverse | GAGGTTTGGCAGGAGCAGAT | ||
| P2 | Forward | CAGAGCACAATGTTCCCCCA | 231 |
| Reverse | CAGTGGCCAGGAACCGTATT | ||
| P3 | Forward | AGAAGCAGAAGACCGAGCAA | 132 |
| Reverse | CCCAAAACTGAACCCCTGGA |
Appendix A.3. Polymerase Chain Reaction (PCR)
References
- Wilkie, A.O.; Morriss-Kay, G.M. Genetics of craniofacial development and malformation. Nat. Rev. Genet. 2021, 2, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Levi, B.; Brugman, S.; Wong, V.W.; Grova, M.; Longaker, M.T.; Wan, D.C. Palatogenesis: Engineering, pathways and pathologies. Organogenesis 2011, 7, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharan, D.; Ramanathan, A. Role of IRF6 Gene in Orofacial Clefting: A Systematic Review. Biomed. Pharmacol. J. 2017, 10, 1097. [Google Scholar] [CrossRef]
- Butali, A.; Mossey, P.A.; Adeyemo, W.L.; Eshete, M.A.; Gaines, L.A.; Even, D.; Braimah, R.O.; Aregbesola, B.S.; Rigdon, J.V.; Emeka, C.I.; et al. Novel IRF6 mutations in families with Van Der Woude syndrome and popliteal pterygium syndrome from sub-Saharan Africa. Mol. Genet. Genom. Med. 2014, 2, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.J.; Standley, J.; Compton, J.; Bale, S.; Schutte, B.C.; Murray, J.C. Comparative analysis of IRF6 variants in families with Van der Woude syndrome and popliteal pterygium syndrome using public whole-exome databases. Genet. Med. 2013, 15, 338–344. [Google Scholar] [CrossRef]
- Tan, E.C.; Lim, E.C.; Lee, S.T. De Novo 2.3 Mb microdeletion of 1q32. 2 involving the Van der Woude Syndrome locus. Mol. Cytogenet. 2013, 6, 1–6. [Google Scholar] [CrossRef]
- Lees, M.M.; Winter, R.M.; Malcolm, S.; Saal, H.M.; Chitty, L. Popliteal pterygium syndrome: A clinical study of three families and report of linkage to the Van der Woude syndrome locus on 1q32. J. Med. Genet. 1999, 36, 888–892. [Google Scholar]
- Tattini, L.; D’Aurizio, R.; Magi, A. Detection of genomic structural variants from next-generation sequencing data. Front. Bioeng. Biotechnol. 2015, 3, 92. [Google Scholar] [CrossRef]
- Sanchis-Juan, A.; Stephens, J.; French, C.E.; Gleadall, N.; Mégy, K.; Penkett, C.; Shamardina, O.; Stirrups, K.; Delon, I.; Dewhurst, E.; et al. Complex structural variants in Mendelian disorders: Identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 2018, 10, 95. [Google Scholar] [CrossRef]
- Carvalho, C.; Ramocki, M.B.; Pehlivan, D.; Franco, L.M.; Gonzaga-Jauregui, C.; Fang, P.; McCall, A.; Pivnick, E.K.; Hines-Dowell, S.; Seaver, L.H.; et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat. Genet. 2011, 43, 1074–1081. [Google Scholar] [CrossRef]
- Bergant, G.; Maver, A.; Peterlin, B. Whole-genome sequencing in diagnostics of selected slovenian undiagnosed patients with rare disorders. Life 2021, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Chaisson, M.J.; Wilson, R.K.; Eichler, E.E. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 2015, 16, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Boycott, K.M.; Vanstone, M.R.; Bulman, D.E.; MacKenzie, A.E. Rare-disease genetics in the era of next-generation sequencing: Discovery to translation. Nat. Rev. Genet. 2013, 14, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, H.; Luo, R.; Wu, H.; Zhu, H.; Li, R.; Cao, H.; Wu, B.; Huang, S.; Shao, H.; et al. Structural variation in two human genomes mapped at single-nucleotide resolution by whole genome de novo assembly. Nat. Biotechnol. 2011, 29, 723–730. [Google Scholar] [CrossRef]
- Fakhro, K.A.; Robay, A.; Rodrigues-Flores, J.L.; Mezey, J.G.; Al-Shakaki, A.A.; Chidiac, O.; Stadler, D.; Malek, J.A.; Imam, A.B.; Sheikh, A.; et al. Point of Care Exome Sequencing Reveals Allelic and Phenotypic Heterogeneity Underlying Mendelian disease in Qatar. Hum. Mol. Genet. 2019, 28, 3970–3981. [Google Scholar] [CrossRef]
- Fakhro, K.A.; Yousri, N.A.; Rodriguez-Flores, J.L.; Robay, A.; Staudt, M.R.; Agosto-Perez, F.; Salit, J.; Malek, J.A.; Suhre, K.; Jayyousi, A.; et al. Copy number variations in the genome of the Qatari population. BMC Genom. 2015, 16, 834. [Google Scholar] [CrossRef]
- Qasim, M.; Shaukat, M. Popliteal pterygium syndrome: A rare entity. APSP J. Case Rep. 2012, 3, 5. [Google Scholar]
- Busche, A.; Hehr, U.; Sieg, P.; Gillessen-Kaesbach, G. Van der Woude and Popliteal Pterygium Syndromes: Broad intrafamilial variability in a three-generation family with mutation in IRF6. Am. J. Med. Genet. Part A 2016, 170, 2404–2407. [Google Scholar] [CrossRef]
- Trappe, K.; Emde, A.K.; Ehrlich, H.C.; Reinert, K. Gustaf: Detecting and correctly classifying SVs in the NGS twilight zone. Bioinformatics 2014, 30, 3484–3490. [Google Scholar] [CrossRef]
- Da’as, S.I.; Aamer, W.; Hasan, W.; Al-Maraghi, A.; Al-Kurbi, A.; Kilani, H.; AlRayahi, J.; Zamel, K.; Stotland, M.A.; Fakhro, K.A. PGAP3 Associated with Hyperphosphatasia with Mental Retardation Plays a Novel Role in Brain Morphogenesis and Neuronal Wiring at Early Development. Cells 2020, 9, 1782. [Google Scholar] [CrossRef]
- Consortium, G.P. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Exome, E.V.S.N.G. Sequencing Project (ESP); Exome Variant Server: Seattle, WA, USA, 2011; Available online: http://evs.gs.washington.edu/EVS/ (accessed on 22 January 2019).
- Jeffares, D.C.; Jolly, C.; Hoti, M.; Speed, D.; Shaw, L.; Rallis, C.; Balloux, F.; Dessimoz, C.; Bähler, J.; Sedlazeck, F.J. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017, 8, 14061. [Google Scholar] [CrossRef]
- Geoffroy, V.; Guignard, T.; Kress, A.; Gaillard, J.B.; Solli-Nowlan, T.; Schalk, A.; Gatinois, V.; Dollfus, H.; Scheidecker, S.; Muller, J. AnnotSV and knotAnnotSV: A web server for human structural variations annotations, ranking and analysis. Nucl. Acids Res. 2021, 49, W21–W28. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Kurbi, A.A.; Aliyev, E.; AlSa’afin, S.; Aamer, W.; Palaniswamy, S.; Al-Maraghi, A.; Kilani, H.; Akil, A.A.-S.; Stotland, M.A.; Fakhro, K.A. A Complex Intrachromosomal Rearrangement Disrupting IRF6 in a Family with Popliteal Pterygium and Van der Woude Syndromes. Genes 2023, 14, 849. https://doi.org/10.3390/genes14040849
Al-Kurbi AA, Aliyev E, AlSa’afin S, Aamer W, Palaniswamy S, Al-Maraghi A, Kilani H, Akil AA-S, Stotland MA, Fakhro KA. A Complex Intrachromosomal Rearrangement Disrupting IRF6 in a Family with Popliteal Pterygium and Van der Woude Syndromes. Genes. 2023; 14(4):849. https://doi.org/10.3390/genes14040849
Chicago/Turabian StyleAl-Kurbi, Alya A., Elbay Aliyev, Sana AlSa’afin, Waleed Aamer, Sasirekha Palaniswamy, Aljazi Al-Maraghi, Houda Kilani, Ammira Al-Shabeeb Akil, Mitchell A. Stotland, and Khalid A. Fakhro. 2023. "A Complex Intrachromosomal Rearrangement Disrupting IRF6 in a Family with Popliteal Pterygium and Van der Woude Syndromes" Genes 14, no. 4: 849. https://doi.org/10.3390/genes14040849
APA StyleAl-Kurbi, A. A., Aliyev, E., AlSa’afin, S., Aamer, W., Palaniswamy, S., Al-Maraghi, A., Kilani, H., Akil, A. A.-S., Stotland, M. A., & Fakhro, K. A. (2023). A Complex Intrachromosomal Rearrangement Disrupting IRF6 in a Family with Popliteal Pterygium and Van der Woude Syndromes. Genes, 14(4), 849. https://doi.org/10.3390/genes14040849





