SLC38A10 Deficiency in Mice Affects Plasma Levels of Threonine and Histidine in Males but Not in Females: A Preliminary Characterization Study of SLC38A10−/− Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Genotyping
2.3. β-Galactosidase Staining
2.4. Blood Plasma Collection
2.4.1. Hormone Measurement in Plasma
2.4.2. Amino Acid Measurements
Standards and Calibration Curve
Amino Acid Extraction
Amino Acid Derivatization with AccQ-Tag
Amino Acids Quantification by LC-ESI-MSMS
2.4.3. Olink Proteomics
2.5. RNA Sequencing
2.6. Tissue Collection, Preparation and RNA Extraction
2.7. cDNA Synthesis and Quantitative Real-Time PCR
2.8. Relative Telomere Length Measurement
2.9. Proteomic Measurement in Brain
2.9.1. Sample Preparation
2.9.2. LC-MS/MS
2.9.3. Qualitative Analysis
2.9.4. Quantitative Analysis
3. Results
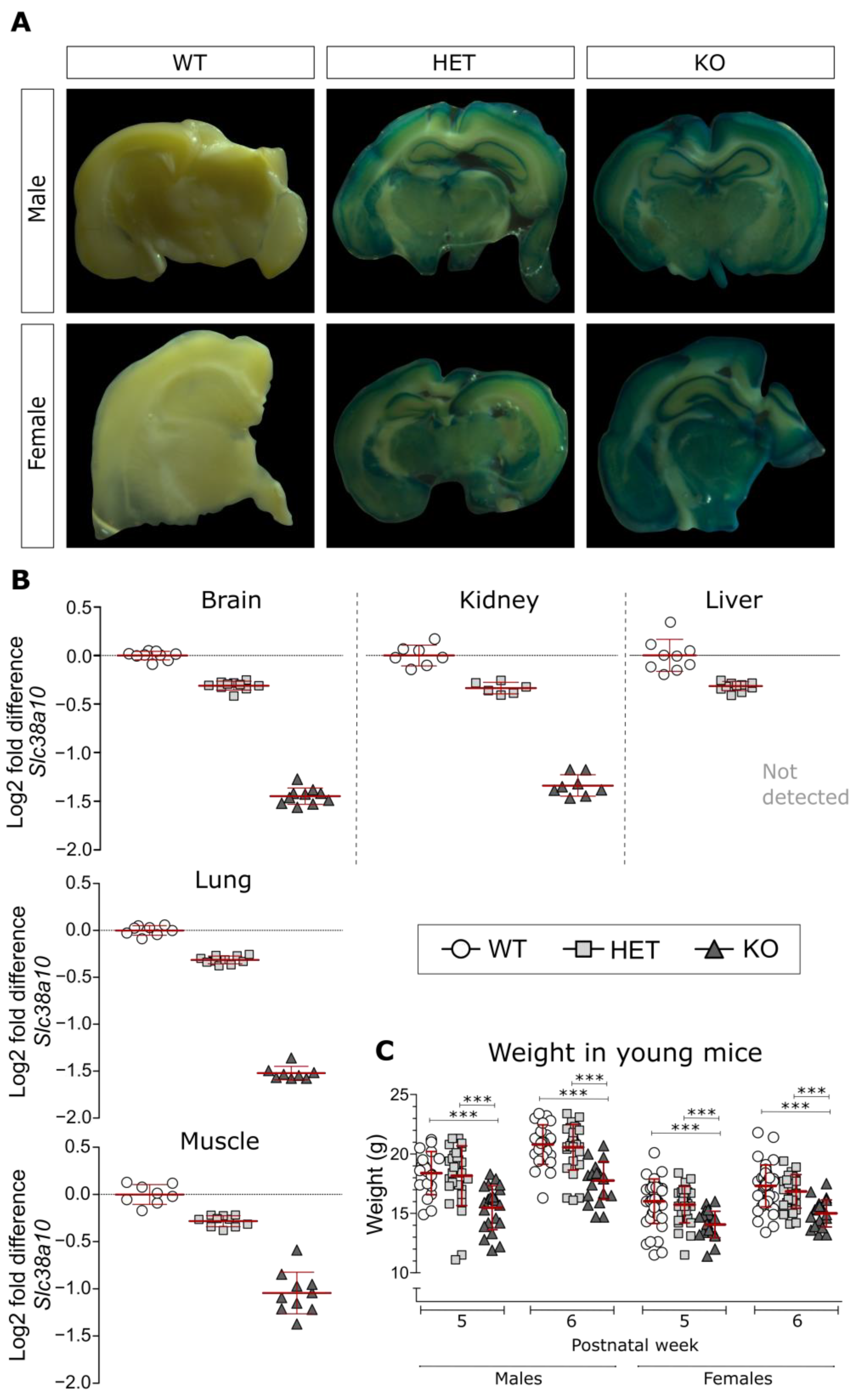
3.1. Analysis of β-Galactosidase and Slc38a10 Expression, as Well as Weight in Young Mice
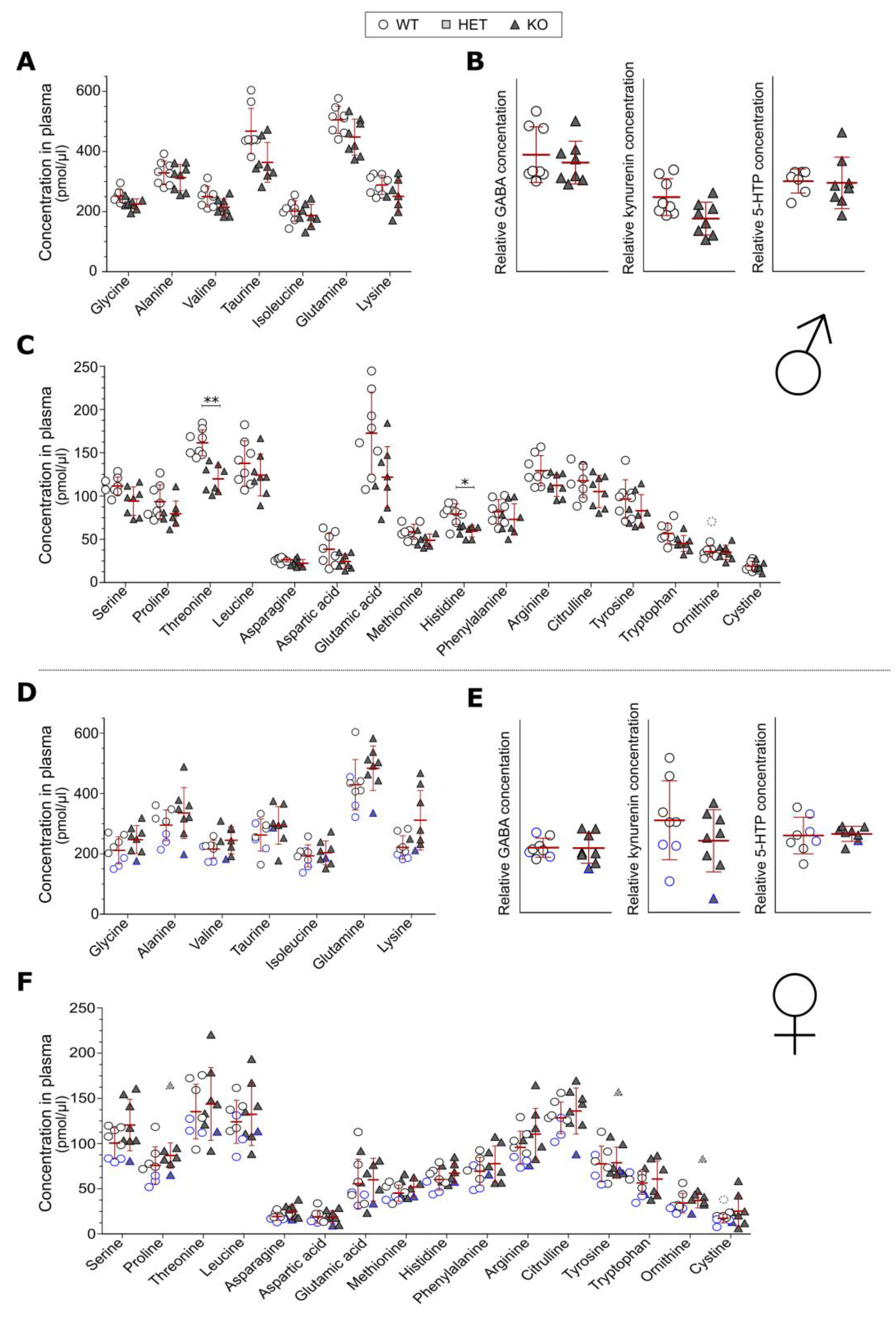
3.2. Blood Plasma Levels of Amino Acids
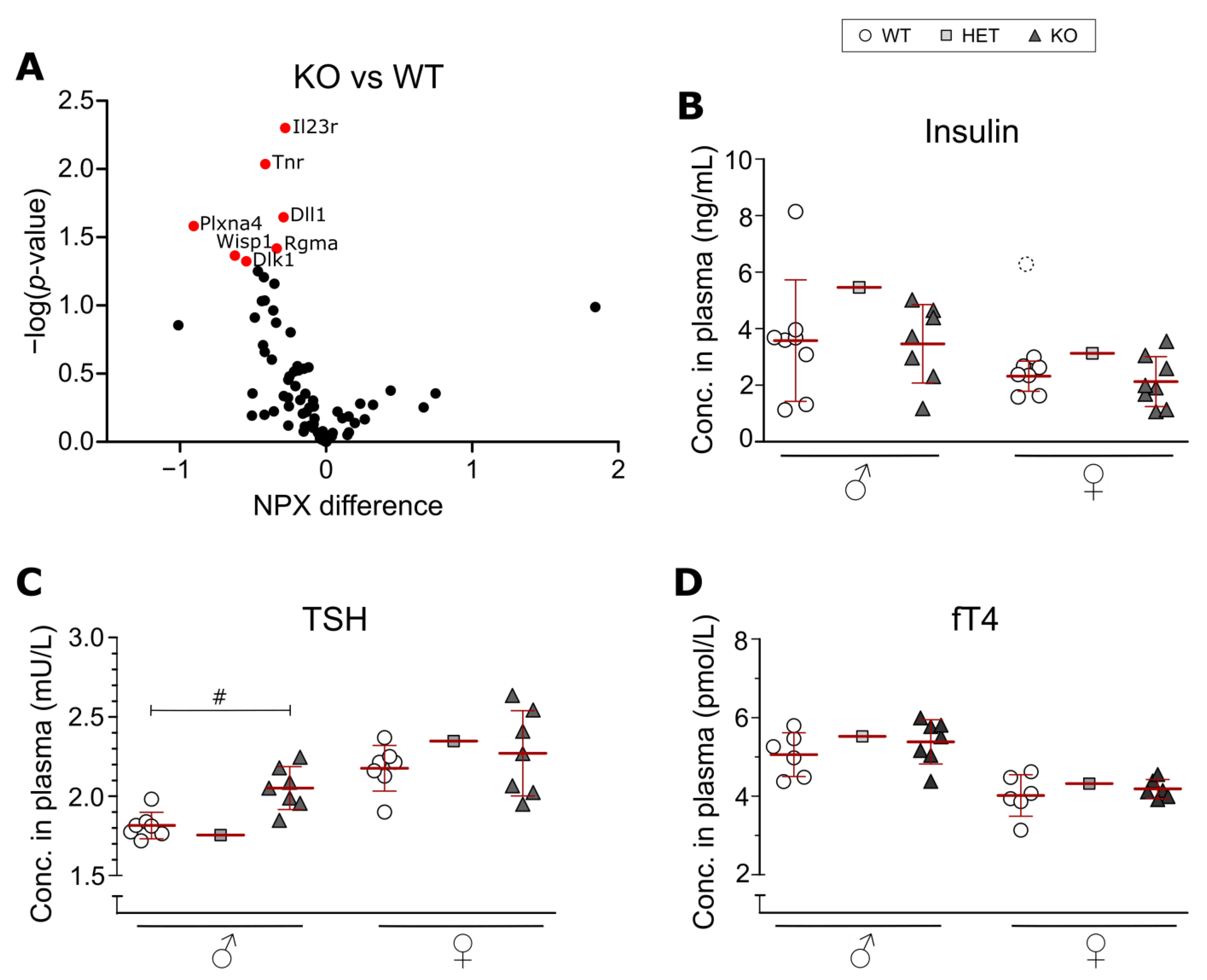
3.3. Olink Proteomics and Metabolic Hormone Measurements in Plasma
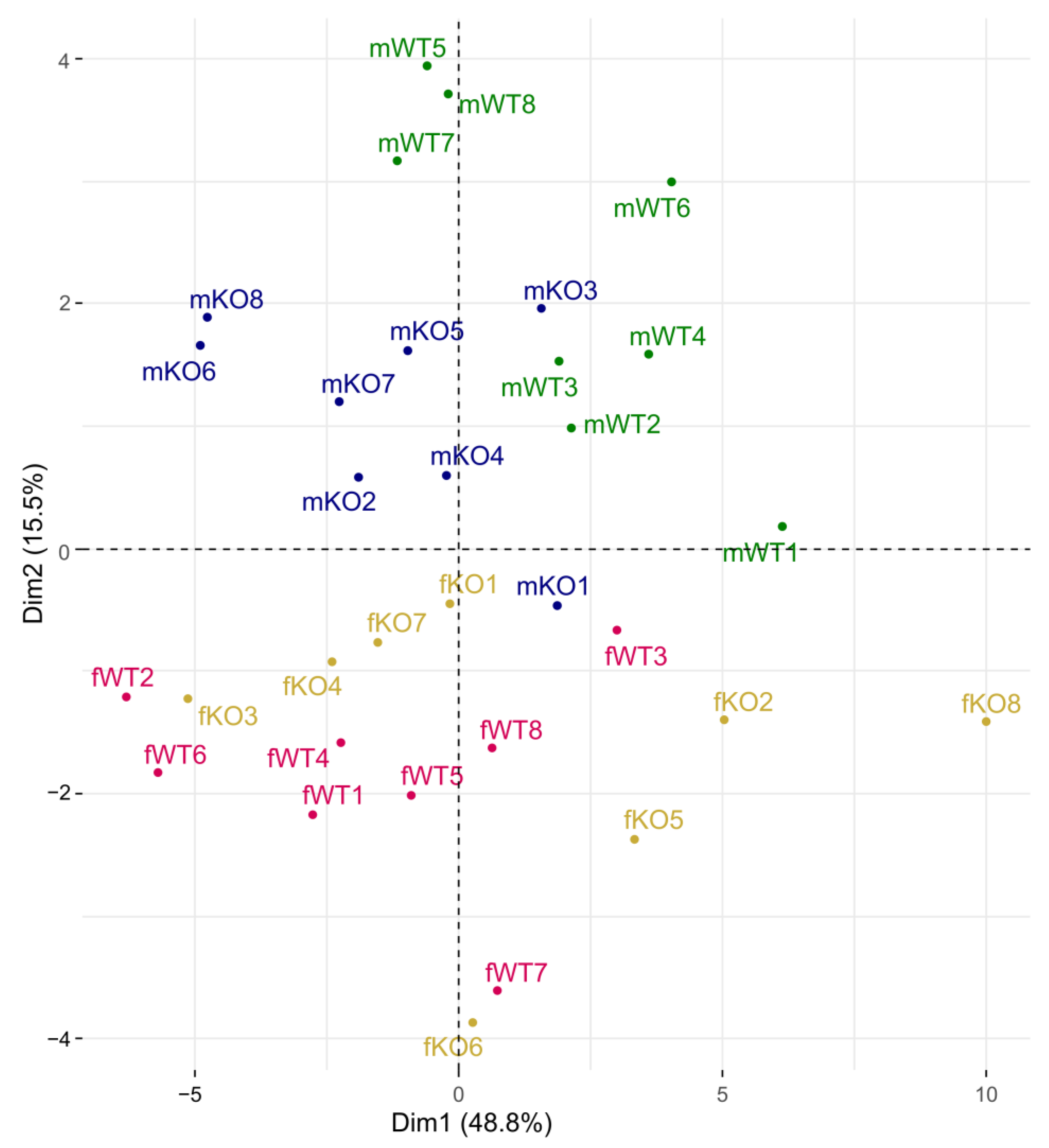
3.4. Transcriptomic Data from Whole Brain
3.5. Gene Expression Analysis in Brain, Kidney, Liver, Lung, and Kidney, as Well as Telomere Length Measurement in Brain
3.6. Brain Proteomics
4. Discussion
4.1. Plasma Levels of Amino Acids Is Affected by SLC38A10
4.2. Effects on Plasma Levels of Insulin, TSH and fT4 in SLC38A10-Deficient Mice
4.3. Whole-Brain Transcriptomics
4.4. Whole-Brain Proteomics
4.5. General Discussion/Limitations of the Current Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hediger, M.A.; Clémençon, B.; Burrier, R.E.; Bruford, E.A. The ABCs of Membrane Transporters in Health and Disease (SLC Series): Introduction. Mol. Aspects Med. 2013, 34, 95–107. [Google Scholar] [CrossRef]
- Gyimesi, G.; Hediger, M.A. SLC66 Lysosomal Amino Acid Transporters in GtoPdb v.2021.2. IUPHAR/BPS Guide to Pharmacology CITE. 2021, Volume 2021. Available online: http://journals.ed.ac.uk/gtopdb-cite/article/view/5736 (accessed on 21 June 2021).
- Hediger, M.A.; Gyimesi, G. SLCtables. Available online: http://slc.bioparadigms.org/ (accessed on 23 August 2020).
- Lin, L.; Yee, S.W.; Kim, R.B.; Giacomini, K.M. SLC Transporters as Therapeutic Targets: Emerging Opportunities. Nat. Rev. Drug Discov. 2015, 14, 543–560. [Google Scholar] [CrossRef]
- César-Razquin, A.; Snijder, B.; Frappier-Brinton, T.; Isserlin, R.; Gyimesi, G.; Bai, X.; Reithmeier, R.A.; Hepworth, D.; Hediger, M.A.; Edwards, A.M.; et al. A Call for Systematic Research on Solute Carriers. Cell 2015, 162, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Bröer, S. The SLC38 Family of Sodium-Amino Acid Co-Transporters. Pflugers Arch. 2014, 466, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, B.; Erickson, J.D. Sodium-Coupled Neutral Amino Acid (System N/A) Transporters of the SLC38 Gene Family. Pflüg. Arch. 2004, 447, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Schiöth, H.B.; Roshanbin, S.; Hägglund, M.G.A.; Fredriksson, R. Evolutionary Origin of Amino Acid Transporter Families SLC32, SLC36 and SLC38 and Physiological, Pathological and Therapeutic Aspects. Mol. Aspects Med. 2013, 34, 571–585. [Google Scholar] [CrossRef]
- Hellsten, S.V.; Hägglund, M.G.; Eriksson, M.M.; Fredriksson, R. The Neuronal and Astrocytic Protein SLC38A10 Transports Glutamine, Glutamate, and Aspartate, Suggesting a Role in Neurotransmission. FEBS Open Bio 2017, 7, 730–746. [Google Scholar] [CrossRef]
- Baird, F.E.; Beattie, K.J.; Hyde, A.R.; Ganapathy, V.; Rennie, M.J.; Taylor, P.M. Bidirectional Substrate Fluxes through the System N (SNAT5) Glutamine Transporter May Determine Net Glutamine Flux in Rat Liver. J. Physiol. 2004, 559, 367–381. [Google Scholar] [CrossRef]
- Blot, A.; Billups, D.; Bjørkmo, M.; Quazi, A.Z.; Uwechue, N.M.; Chaudhry, F.A.; Billups, B. Functional Expression of Two System A Glutamine Transporter Isoforms in Rat Auditory Brainstem Neurons. Neuroscience 2009, 164, 998–1008. [Google Scholar] [CrossRef]
- Boulland, J.-L.; Osen, K.K.; Levy, L.M.; Danbolt, N.C.; Edwards, R.H.; Storm-Mathisen, J.; Chaudhry, F.A. Cell-Specific Expression of the Glutamine Transporter SN1 Suggests Differences in Dependence on the Glutamine Cycle. Eur. J. Neurosci. 2002, 15, 1615–1631. [Google Scholar] [CrossRef]
- Chaudhry, F.A.; Reimer, R.J.; Krizaj, D.; Barber, D.; Storm-Mathisen, J.; Copenhagen, D.R.; Edwards, R.H. Molecular Analysis of System N Suggests Novel Physiological Roles in Nitrogen Metabolism and Synaptic Transmission. Cell 1999, 99, 769–780. [Google Scholar] [CrossRef]
- Cubelos, B.; González-González, I.M.; Giménez, C.; Zafra, F. Amino Acid Transporter SNAT5 Localizes to Glial Cells in the Rat Brain. Glia 2005, 49, 230–244. [Google Scholar] [CrossRef]
- Gandasi, N.R.; Arapi, V.; Mickael, M.E.; Belekar, P.A.; Granlund, L.; Kothegala, L.; Fredriksson, R.; Bagchi, S. Glutamine Uptake via SNAT6 and Caveolin Regulates Glutamine-Glutamate Cycle. Int. J. Mol. Sci. 2021, 22, 1167. [Google Scholar] [CrossRef]
- Hägglund, M.G.A.; Hellsten, S.V.; Bagchi, S.; Philippot, G.; Löfqvist, E.; Nilsson, V.C.O.; Almkvist, I.; Karlsson, E.; Sreedharan, S.; Tafreshiha, A.; et al. Transport of L-Glutamine, L-Alanine, L-Arginine and L-Histidine by the Neuron-Specific Slc38a8 (SNAT8) in CNS. J. Mol. Biol. 2015, 427, 1495–1512. [Google Scholar] [CrossRef]
- Hägglund, M.G.A.; Sreedharan, S.; Nilsson, V.C.O.; Shaik, J.H.A.; Almkvist, I.M.; Bäcklin, S.; Wrange, O.; Fredriksson, R. Identification of SLC38A7 (SNAT7) Protein as a Glutamine Transporter Expressed in Neurons. J. Biol. Chem. 2011, 286, 20500–20511. [Google Scholar] [CrossRef]
- Hatanaka, T.; Huang, W.; Ling, R.; Prasad, P.D.; Sugawara, M.; Leibach, F.H.; Ganapathy, V. Evidence for the Transport of Neutral as Well as Cationic Amino Acids by ATA3, a Novel and Liver-Specific Subtype of Amino Acid Transport System A. Biochim. Biophys. Acta 2001, 1510, 10–17. [Google Scholar] [CrossRef]
- Varoqui, H.; Erickson, J.D. Selective Up-Regulation of System a Transporter MRNA in Diabetic Liver. Biochem. Biophys. Res. Commun. 2002, 290, 903–908. [Google Scholar] [CrossRef]
- Sundberg, B.E.; Wååg, E.; Jacobsson, J.A.; Stephansson, O.; Rumaks, J.; Svirskis, S.; Alsiö, J.; Roman, E.; Ebendal, T.; Klusa, V.; et al. The Evolutionary History and Tissue Mapping of Amino Acid Transporters Belonging to Solute Carrier Families SLC32, SLC36, and SLC38. J. Mol. Neurosci. MN 2008, 35, 179–193. [Google Scholar] [CrossRef]
- Cedernaes, J.; Olszewski, P.K.; Almén, M.S.; Stephansson, O.; Levine, A.S.; Fredriksson, R.; Nylander, O.; Schiöth, H.B. Comprehensive Analysis of Localization of 78 Solute Carrier Genes throughout the Subsections of the Rat Gastrointestinal Tract. Biochem. Biophys. Res. Commun. 2011, 411, 702–707. [Google Scholar] [CrossRef]
- Hellsten, S.V.; Tripathi, R.; Ceder, M.M.; Fredriksson, R. Nutritional Stress Induced by Amino Acid Starvation Results in Changes for Slc38 Transporters in Immortalized Hypothalamic Neuronal Cells and Primary Cortex Cells. Front. Mol. Biosci. 2018, 5, 45. [Google Scholar] [CrossRef]
- Tripathi, R.; Hosseini, K.; Arapi, V.; Fredriksson, R.; Bagchi, S. SLC38A10 (SNAT10) Is Located in ER and Golgi Compartments and Has a Role in Regulating Nascent Protein Synthesis. Int. J. Mol. Sci. 2019, 20, 6265. [Google Scholar] [CrossRef] [PubMed]
- SLC38A10 Protein Expression Summary—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000157637-SLC38A10 (accessed on 19 February 2023).
- Jung, J.; Genau, H.M.; Behrends, C. Amino Acid-Dependent MTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol. Cell. Biol. 2015, 35, 2479–2494. [Google Scholar] [CrossRef]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; de Araújo, M.E.G.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 Is a Component of the Lysosomal Amino Acid-Sensing Machinery That Controls MTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tsun, Z.-Y.; Wolfson, R.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W.; et al. The Amino Acid Transporter SLC38A9 Is a Key Component of a Lysosomal Membrane Complex That Signals Arginine Sufficiency to MTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Wyant, G.A.; Abu-Remaileh, M.; Wolfson, R.L.; Chen, W.W.; Freinkman, E.; Danai, L.V.; Vander Heiden, M.G.; Sabatini, D.M. MTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell 2017, 171, 642–654.e12. [Google Scholar] [CrossRef]
- Evans, K.; Nasim, Z.; Brown, J.; Butler, H.; Kauser, S.; Varoqui, H.; Erickson, J.D.; Herbert, T.P.; Bevington, A. Acidosis-Sensing Glutamine Pump SNAT2 Determines Amino Acid Levels and Mammalian Target of Rapamycin Signalling to Protein Synthesis in L6 Muscle Cells. J. Am. Soc. Nephrol. 2007, 18, 1426–1436. [Google Scholar] [CrossRef]
- Pinilla, J.; Aledo, J.C.; Cwiklinski, E.; Hyde, R.; Taylor, P.M.; Hundal, H.S. SNAT2 Transceptor Signalling via MTOR: A Role in Cell Growth and Proliferation? Front. Biosci. Elite Ed. 2011, 3, 1289–1299. [Google Scholar] [CrossRef]
- Tripathi, R.; Aggarwal, T.; Fredriksson, R. SLC38A10 Transporter Plays a Role in Cell Survival Under Oxidative Stress and Glutamate Toxicity. Front. Mol. Biosci. 2021, 8, 671865. [Google Scholar] [CrossRef]
- Fan, S.-J.; Goberdhan, D.C.I. PATs and SNATs: Amino Acid Sensors in Disguise. Front. Pharmacol. 2018, 9, 640. [Google Scholar] [CrossRef]
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-Throughput Discovery of Novel Developmental Phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef]
- Lindberg, F.A.; Nordenankar, K.; Fredriksson, R. SLC38A10 Knockout Mice Display a Decreased Body Weight and an Increased Risk-Taking Behavior in the Open Field Test. Front. Behav. Neurosci. 2022, 16, 840987. [Google Scholar] [CrossRef]
- Skarnes, W.C.; Rosen, B.; West, A.P.; Koutsourakis, M.; Bushell, W.; Iyer, V.; Mujica, A.O.; Thomas, M.; Harrow, J.; Cox, T.; et al. A Conditional Knockout Resource for the Genome-Wide Study of Mouse Gene Function. Nature 2011, 474, 337–342. [Google Scholar] [CrossRef]
- Testa, G.; Schaft, J.; van der Hoeven, F.; Glaser, S.; Anastassiadis, K.; Zhang, Y.; Hermann, T.; Stremmel, W.; Stewart, A.F. A Reliable LacZ Expression Reporter Cassette for Multipurpose, Knockout-First Alleles. Genesis 2004, 38, 151–158. [Google Scholar] [CrossRef]
- Slc38a10—Solute Carrier Family 38, Member 10|International Mouse Phenotyping Consortium. Available online: https://www.mousephenotype.org/data/genes/MGI:1919305 (accessed on 1 September 2020).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinform. Oxf. Engl. 2013, 29, 15–21. [Google Scholar] [CrossRef]
- R: Documentation. Available online: https://www.r-project.org/other-docs.html (accessed on 2 June 2021).
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential Analysis of Gene Regulation at Transcript Resolution with RNA-Seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Roberts, A.; Pimentel, H.; Trapnell, C.; Pachter, L. Identification of Novel Transcripts in Annotated Genomes Using RNA-Seq. Bioinforma. Oxf. Engl. 2011, 27, 2325–2329. [Google Scholar] [CrossRef]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq Expression Estimates by Correcting for Fragment Bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Cantó, C.; Jeninga, E.H.; Andreux, P.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The Metabolic Footprint of Aging in Mice. Sci. Rep. 2011, 1, 134. [Google Scholar] [CrossRef]
- Tripathi, R.; Aggarwal, T.; Lindberg, F.A.; Klemm, A.H.; Fredriksson, R. SLC38A10 Regulate Glutamate Homeostasis and Modulate the AKT/TSC2/MTOR Pathway in Mouse Primary Cortex Cells. Front. Cell Dev. Biol. 2022, 10, 854397. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Tan, P.; Ma, N.; Ma, X. Physiological Functions of Threonine in Animals: Beyond Nutrition Metabolism. Nutrients 2021, 13, 2592. [Google Scholar] [CrossRef] [PubMed]
- Boehm, G.; Cervantes, H.; Georgi, G.; Jelinek, J.; Sawatzki, G.; Wermuth, B.; Colombo, J.-P. Effect of Increasing Dietary Threonine Intakes on Amino Acid Metabolism of the Central Nervous System and Peripheral Tissues in Growing Rats. Pediatr. Res. 1998, 44, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Holeček, M. Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients 2020, 12, 848. [Google Scholar] [CrossRef] [PubMed]
- Weiner, C.P.; Lizasoain, I.; Baylis, S.A.; Knowles, R.G.; Charles, I.G.; Moncada, S. Induction of Calcium-Dependent Nitric Oxide Synthases by Sex Hormones. Proc. Natl. Acad. Sci. USA 1994, 91, 5212–5216. [Google Scholar] [CrossRef]
- Tipton, K.D. Gender Differences in Protein Metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2001, 4, 493–498. [Google Scholar] [CrossRef]
- Darst, B.F.; Koscik, R.L.; Hogan, K.J.; Johnson, S.C.; Engelman, C.D. Longitudinal Plasma Metabolomics of Aging and Sex. Aging 2019, 11, 1262–1282. [Google Scholar] [CrossRef]
- Bröer, S.; Bröer, A. Amino Acid Homeostasis and Signalling in Mammalian Cells and Organisms. Biochem. J. 2017, 474, 1935–1963. [Google Scholar] [CrossRef]
- Bassett, J.H.D.; Gogakos, A.; White, J.K.; Evans, H.; Jacques, R.M.; van der Spek, A.H.; Ramirez-Solis, R.; Ryder, E.; Sunter, D.; Boyde, A.; et al. Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength. PLoS Genet. 2012, 8, e1002858. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Mohan, S. Role and Mechanisms of Actions of Thyroid Hormone on the Skeletal Development. Bone Res. 2013, 1, 146–161. [Google Scholar] [CrossRef]
- Koulouri, O.; Gurnell, M. How to Interpret Thyroid Function Tests. Clin. Med. 2013, 13, 282–286. [Google Scholar] [CrossRef]
- Nakajima, Y.; Yamada, M.; Taguchi, R.; Shibusawa, N.; Ozawa, A.; Tomaru, T.; Hashimoto, K.; Saito, T.; Tsuchiya, T.; Okada, S.; et al. NR4A1 (Nur77) Mediates Thyrotropin-Releasing Hormone-Induced Stimulation of Transcription of the Thyrotropin β Gene: Analysis of TRH Knockout Mice. PLoS ONE 2012, 7, e40437. [Google Scholar] [CrossRef]
- Glotzer, M. The 3Ms of Central Spindle Assembly: Microtubules, Motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009, 10, 9–20. [Google Scholar] [CrossRef]
- Bian, T.; Zheng, M.; Jiang, D.; Liu, J.; Sun, H.; Li, X.; Liu, L.; Zhang, J.; Liu, Y. Prognostic Biomarker TUBA1C Is Correlated to Immune Cell Infiltration in the Tumor Microenvironment of Lung Adenocarcinoma. Cancer Cell Int. 2021, 21, 144. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef]
- Bonnefoy, S.; Watson, C.M.; Kernohan, K.D.; Lemos, M.; Hutchinson, S.; Poulter, J.A.; Crinnion, L.A.; Berry, I.; Simmonds, J.; Vasudevan, P.; et al. Biallelic Mutations in LRRC56, Encoding a Protein Associated with Intraflagellar Transport, Cause Mucociliary Clearance and Laterality Defects. Am. J. Hum. Genet. 2018, 103, 727–739. [Google Scholar] [CrossRef]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic SnoRNAs: A Paradigm for Gene Expression Flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef]
- Ding, F.; Li, H.H.; Zhang, S.; Solomon, N.M.; Camper, S.A.; Cohen, P.; Francke, U. SnoRNA Snord116 (Pwcr1/MBII-85) Deletion Causes Growth Deficiency and Hyperphagia in Mice. PLoS ONE 2008, 3, e1709. [Google Scholar] [CrossRef]
- Coulson, R.L.; Yasui, D.H.; Dunaway, K.W.; Laufer, B.I.; Vogel Ciernia, A.; Zhu, Y.; Mordaunt, C.E.; Totah, T.S.; LaSalle, J.M. Snord116-Dependent Diurnal Rhythm of DNA Methylation in Mouse Cortex. Nat. Commun. 2018, 9, 1616. [Google Scholar] [CrossRef]
- Buechler, C.; Bodzioch, M.; Bared, S.M.; Sigruener, A.; Boettcher, A.; Lapicka-Bodzioch, K.; Aslanidis, C.; Duong, C.Q.; Grandl, M.; Langmann, T.; et al. Expression Pattern and Raft Association of NIPSNAP3 and NIPSNAP4, Highly Homologous Proteins Encoded by Genes in Close Proximity to the ATP-Binding Cassette Transporter A1. Genomics 2004, 83, 1116–1124. [Google Scholar] [CrossRef]
- The UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [CrossRef] [PubMed]
- El-Brolosy, M.A.; Stainier, D.Y.R. Genetic Compensation: A Phenomenon in Search of Mechanisms. PLoS Genet. 2017, 13, e1006780. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
| Gene: | Primer (5′-3′): |
|---|---|
| H3c | Forward: ccttgtgggtctgtttga; Reverse: cagttggatgtccttggg |
| Actb | Forward: ccttcttgggtatggaatcctgtg; Reverse: cagcactgtgttggcatagagg |
| Tubb4b | Forward: agtgctcctcttctacag; Reverse: tatctccgtggtaagtgc |
| mTOR | Forward: acatttgaagaagcagag; Reverse: tgatctcctccatctctt |
| Rps6kb1 | Forward: actagtgtgaacagagggcc; Reverse: ttcctccagaatgttccgct |
| Slc38a1 | Forward: tcacgagagtcacgcagagatg; Reverse: gagcagtatgagaacaagcgaagc |
| Slc38a2 | Forward: tctactcgctggttcttc; Reverse: aataaacttgtcacttccctta |
| Slc38a3 | Forward: atatttgggatcattggt; Reverse: catgattcggaagtagaa |
| Slc38a4 | Forward: agaacatcagacaaagac; Reverse: ttagtgttggtgtagagt |
| Slc38a5 | Forward: tggaggtgtctggtctctaataac; Reverse: ggcagtgaggcaactctaagg |
| Slc38a7 | Forward: tagccattgcggtctata; Reverse: gctccttcgacatcacag |
| Slc38a8 | Forward: cgtggtgactcgggacag; Reverse: ctacaagccagggacactaagg |
| Slc38a9 | Forward: tgctgtcattgctgtaat; Reverse: actgagaagaaccatcct |
| Slc38a10 | Forward: tggtgaaggctccgaagaaagg; Reverse: acttggcttgggtctgaactgg |
| Slc38a11 | Forward: actttcaattcggaacct; Reverse: catcagtgctaatcttgtg |
| log2 | Adjusted | Gene | |
|---|---|---|---|
| Fold Change | p-Value | ||
| ENSMUSG00000061306 | 4.261195835 | 4.92 × 10-210 | Slc38a10 |
| ENSMUSG00000110831 | 4.475565312 | 6.32 × 10-9 | Gm48159 |
| ENSMUSG00000023034 | 0.84989094 | 0.000108 | Nr4a1 |
| ENSMUSG00000043091 | −0.70844378 | 0.004278 | Tba1c |
| ENSMUSG00000038637 | −1.25011731 | 0.006453 | Lrrc56 |
| ENSMUSG00000064372 | −1.42276608 | 0.007218 | mt-Tp |
| ENSMUSG00000073940 | −0.77138591 | 0.008367 | Hbb-bt |
| ENSMUSG00000096634 | −1.36720258 | 0.009128 | Snord116/9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindberg, F.A.; Nordenankar, K.; Forsberg, E.C.; Fredriksson, R. SLC38A10 Deficiency in Mice Affects Plasma Levels of Threonine and Histidine in Males but Not in Females: A Preliminary Characterization Study of SLC38A10−/− Mice. Genes 2023, 14, 835. https://doi.org/10.3390/genes14040835
Lindberg FA, Nordenankar K, Forsberg EC, Fredriksson R. SLC38A10 Deficiency in Mice Affects Plasma Levels of Threonine and Histidine in Males but Not in Females: A Preliminary Characterization Study of SLC38A10−/− Mice. Genes. 2023; 14(4):835. https://doi.org/10.3390/genes14040835
Chicago/Turabian StyleLindberg, Frida A., Karin Nordenankar, Erica C. Forsberg, and Robert Fredriksson. 2023. "SLC38A10 Deficiency in Mice Affects Plasma Levels of Threonine and Histidine in Males but Not in Females: A Preliminary Characterization Study of SLC38A10−/− Mice" Genes 14, no. 4: 835. https://doi.org/10.3390/genes14040835
APA StyleLindberg, F. A., Nordenankar, K., Forsberg, E. C., & Fredriksson, R. (2023). SLC38A10 Deficiency in Mice Affects Plasma Levels of Threonine and Histidine in Males but Not in Females: A Preliminary Characterization Study of SLC38A10−/− Mice. Genes, 14(4), 835. https://doi.org/10.3390/genes14040835






