Development of Improved DNA Collection and Extraction Methods for Handled Documents
Abstract
1. Introduction
2. Materials and Methods
2.1. Dry/Dry Flocked Swab Technique
2.2. Wet/Dry Flocked Swab Technique
2.3. Direct Cutting Technique
2.4. Vacuum Swab Technique
2.4.1. Glass Pasteur Pipette
2.4.2. Plastic Pipette Tip
2.5. Extraction, Quantitation, Amplification
2.6. Stability Study
3. Results
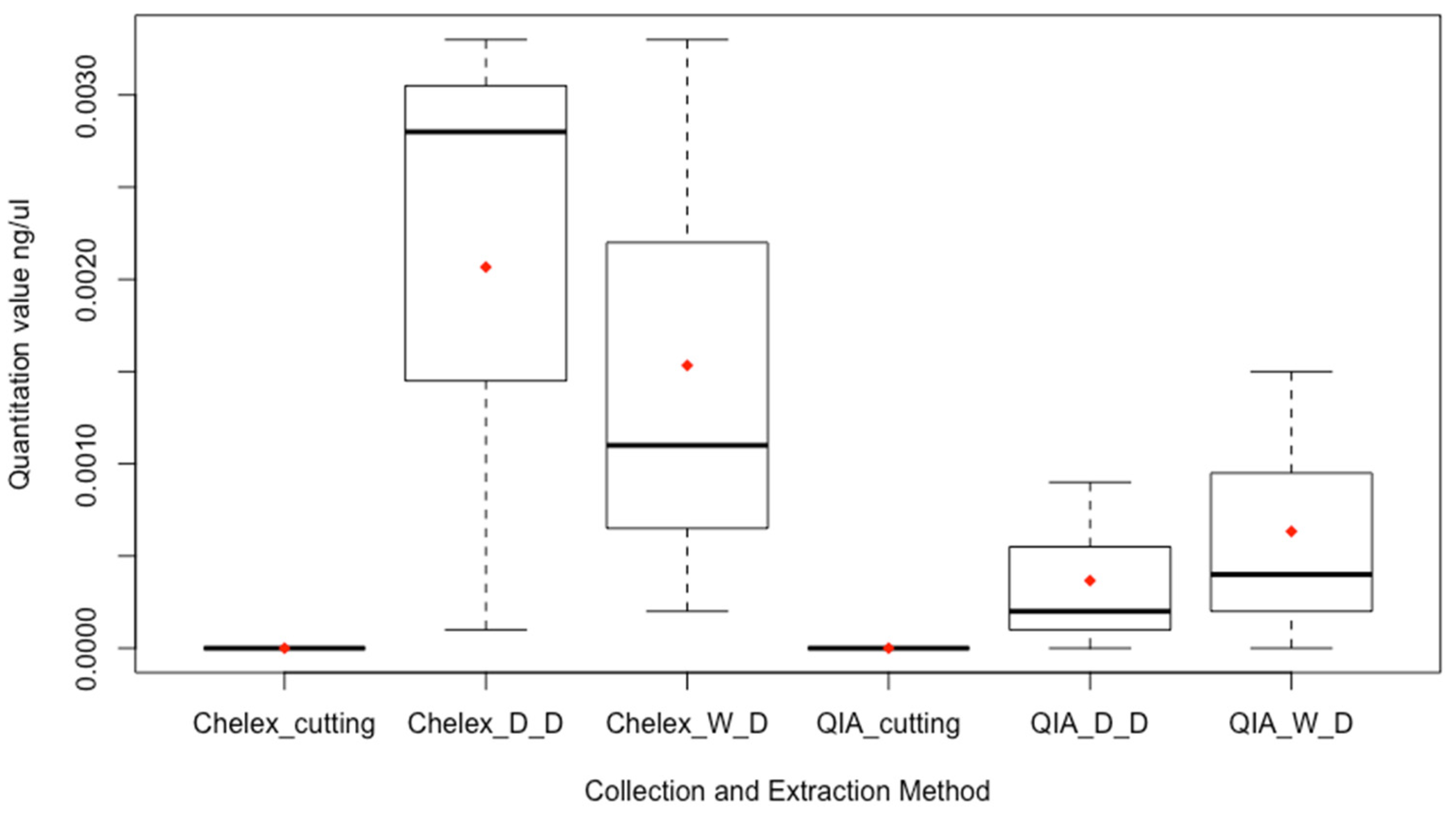
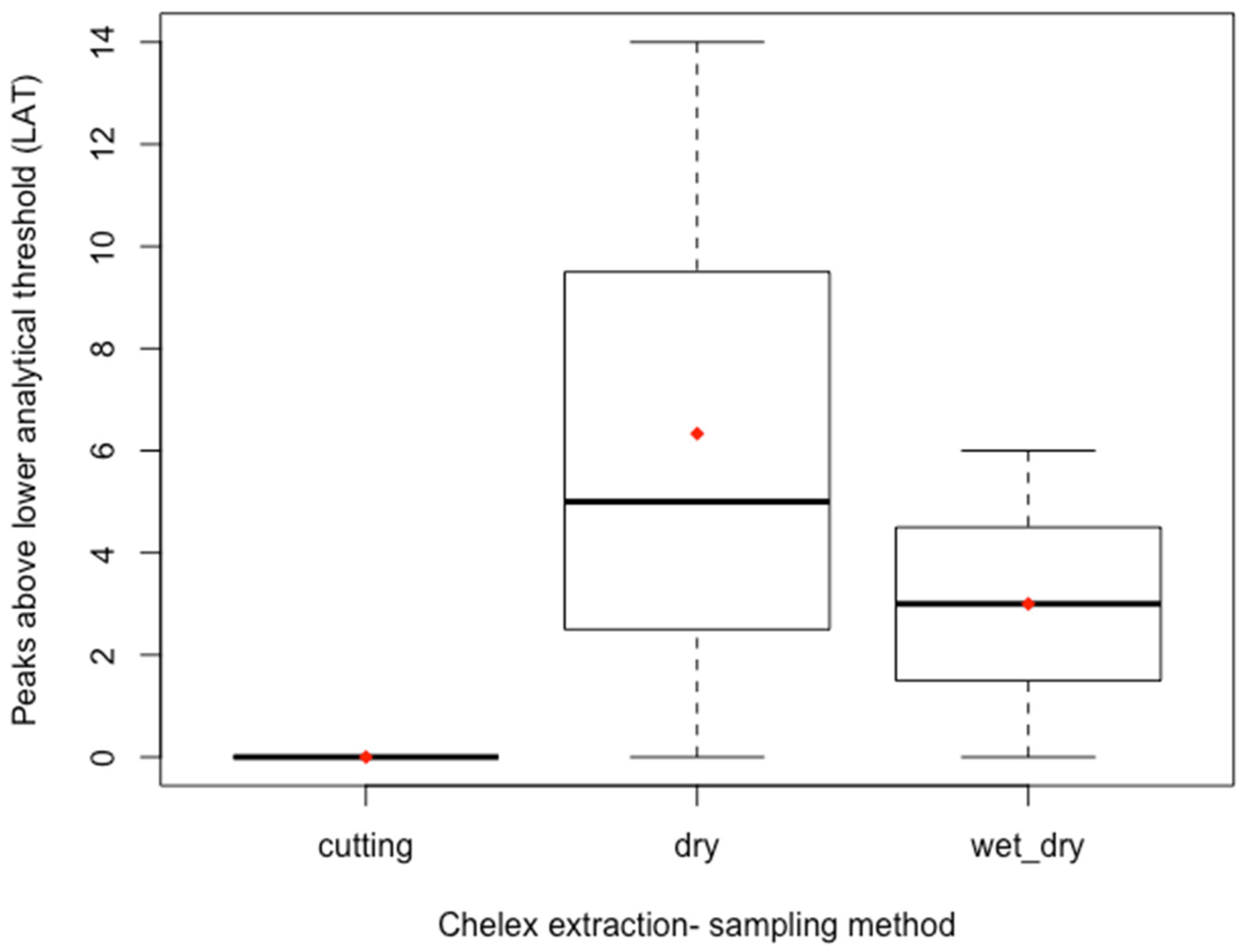
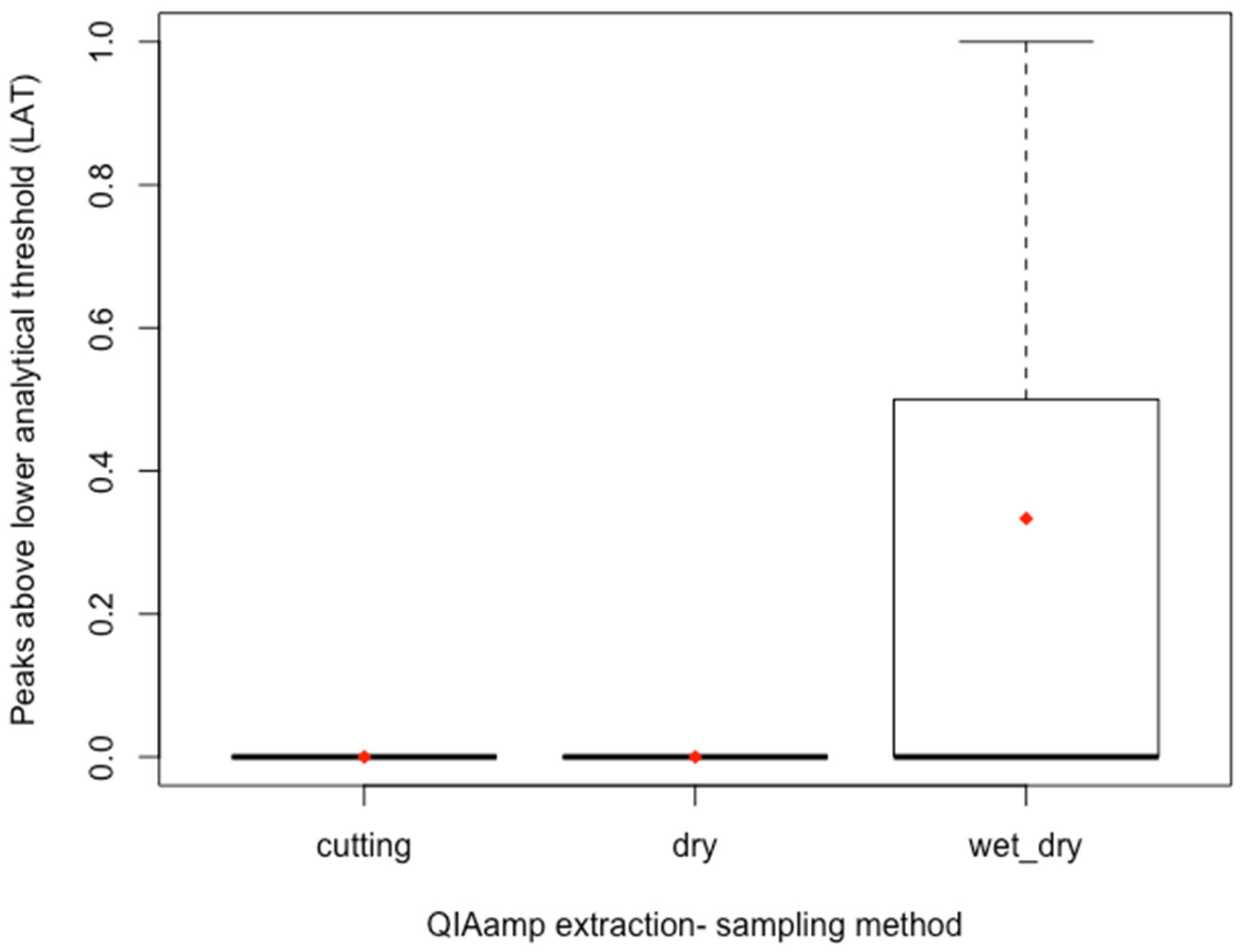
3.1. Comparison of Sampling Technique
3.2. Vacuum Swab Technique
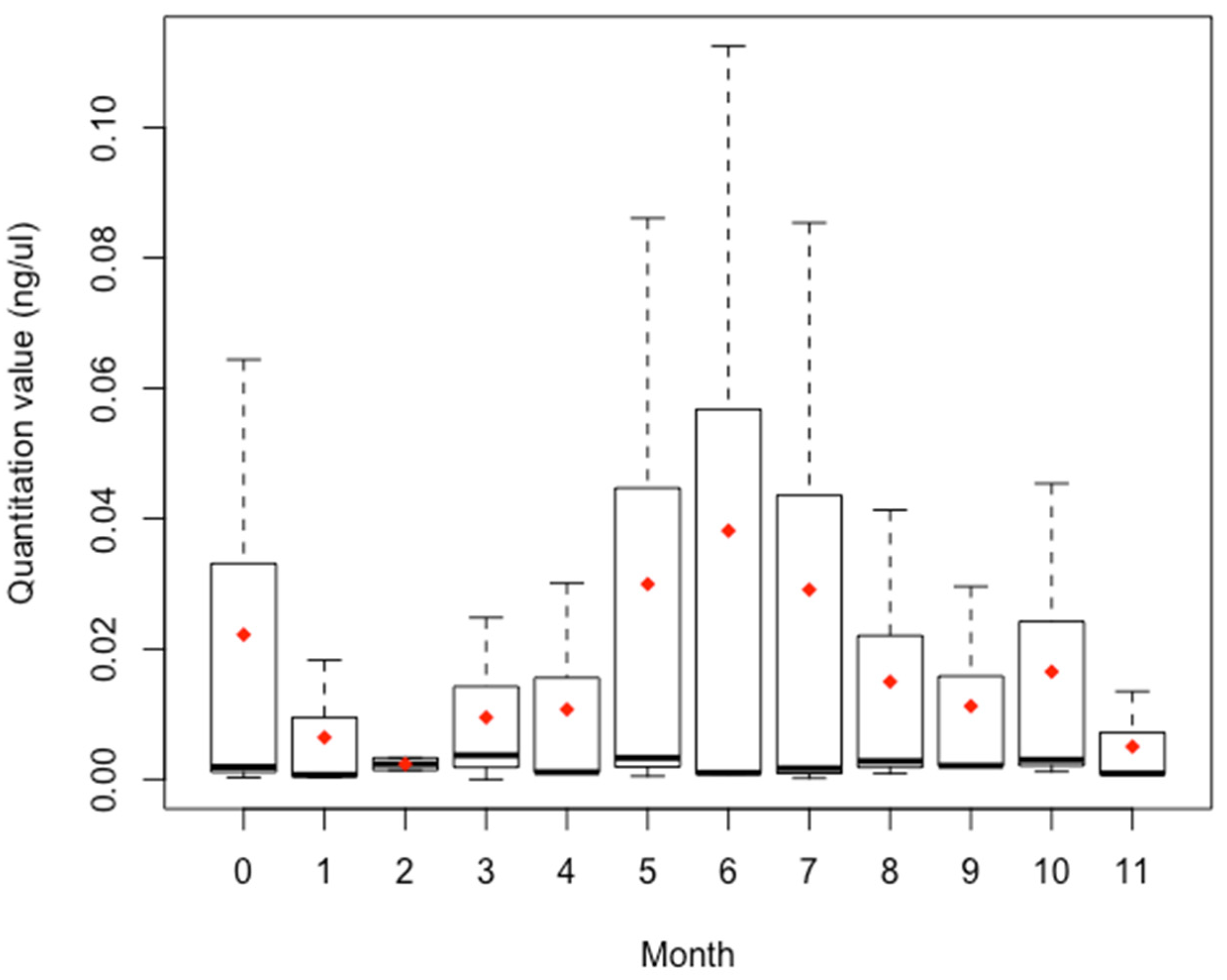
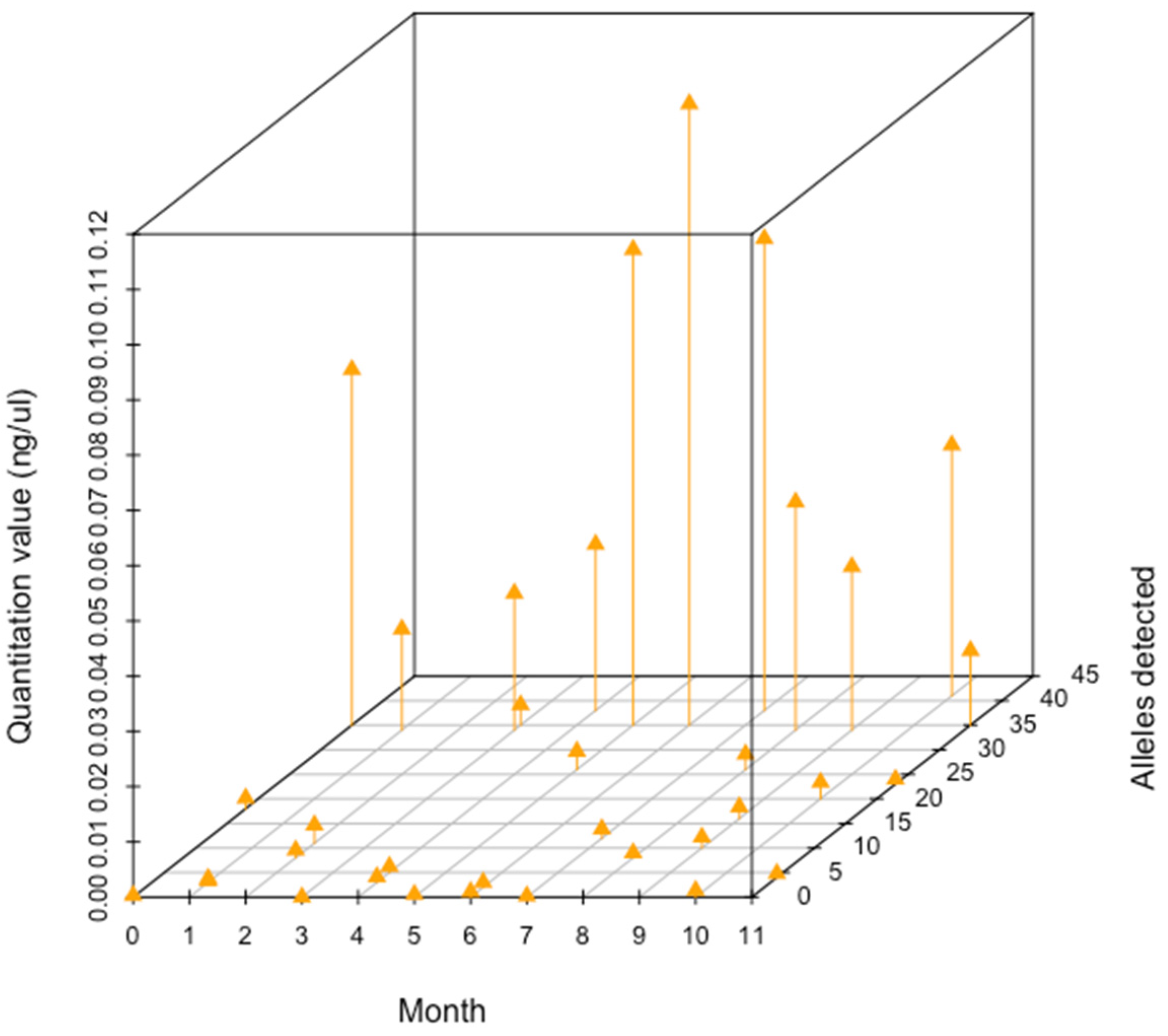
3.3. Stability Study
4. Discussion
4.1. Direct Cutting and Swabbing Study
4.2. Vacuum Swab Study
4.3. Stability Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lowe, A.; Murray, C.; Whitaker, J.; Tully, G.; Gill, P. The Propensity of Individuals to Deposit DNA and Secondary Transfer of Low Level DNA from Individuals to Inert Surfaces. Forensic Sci. Int. 2002, 129, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Balogh, M.K.; Burger, J.; Bender, K.; Schneider, P.M.; Alt, K.W. STR Genotyping and MtDNA Sequencing of Latent Fingerprint on Paper. Forensic Sci. Int. 2003, 137, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Oleiwi, A.A.; Morris, M.R.; Schmerer, W.M.; Sutton, R. The Relative DNA-Shedding Propensity of the Palm and Finger Surfaces. Sci. Justice 2015, 55, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Quinones, I.; Daniel, B. Cell Free DNA as a Component of Forensic Evidence Recovered from Touched Surfaces. Forensic Sci. Int. Genet. 2012, 6, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Jansson, L.; Swensson, M.; Gifvars, E.; Hedell, R.; Forsberg, C.; Ansell, R.; Hedman, J. Individual Shedder Status and the Origin of Touch DNA. Forensic Sci. Int. Genet. 2022, 56, 102626. [Google Scholar] [CrossRef] [PubMed]
- Kanokwongnuwut, P.; Kirkbride, P.; Linacre, A. Speed of accumulation of DNA in a fingermark. Aust. J. Forensic Sci. 2022, 52, 293–302. [Google Scholar] [CrossRef]
- Parsons, L.; Sharfe, G.; Vintiner, S. DNA Analysis and Document Examination: The Impact of Each Technique on Respective Analyses. J. Forensic Sci. 2016, 61, 26–34. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, P.; Hopkins, C.; Springer, E.; Prinz, M. Non-Destructive DNA Recovery from Handwritten Documents Using a Dry Vacuum Technique. J. Forensic Sci. 2021, 66, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Comte, J.; Baechler, S.; Gervaix, J.; Lock, E.; Milon, M.P.; Delémont, O.; Castella, V. Touch DNA Collection—Performance of Four Different Swabs. Forensic Sci. Int. Genet. 2019, 43, 102113. [Google Scholar] [CrossRef] [PubMed]
- Bruijns, B.B.; Tiggelaar, R.M.; Gardeniers, H. The Extraction and Recovery Efficiency of Pure DNA for Different Types of Swabs. J. Forensic Sci. 2018, 63, 1492–1499. [Google Scholar] [CrossRef] [PubMed]
- Sewell, J.; Quinones, I.; Ames, C.; Multaney, B.; Curtis, S.; Seeboruth, H.; Moore, S.; Daniel, B. Recovery of DNA and Fingerprints from Touched Documents. Forensic Sci. Int. Genet. 2008, 2, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Plaza, D.T.; Mealy, J.L.; Lane, J.N.; Parsons, M.N.; Bathrick, A.S.; Slack, D.P. ESDA®-Lite Collection of DNA from Latent Fingerprints on Documents. Forensic Sci. Int. Genet. 2015, 16, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, C.; Jansson, L.; Ansell, R.; Hedman, J. High-Throughput DNA Extraction of Forensic Adhesive Tapes. Forensic Sci. Int. Genet. 2016, 24, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Thermo Fisher Scientific. QuantifilerTM HP and Trio DNA QuantificationKits. 2017. Available online: https://tools.thermofisher.com/content/sfs/manuals/4485354.pdf (accessed on 11 February 2023).
- Ludeman, M.J.; Zhong, C.; Mulero, J.J.; Lagacé, R.E.; Hennessy, L.K.; Short, M.L.; Wang, D.Y. Developmental Validation of GlobalFilerTM PCR Amplification Kit: A 6-Dye Multiplex Assay Designed for Amplification of Casework Samples. Int. J. Legal Med. 2018, 132, 1555–1573. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A. Scientific Development of an Integrated Workflow for Latent Print, Questioned Document, and DNA Processing of Paper Evidence. 2022. Available online: https://academicworks.cuny.edu/gc_etds/4678Discoveradditionalworksat:https://academicworks.cuny.edu (accessed on 11 February 2023).
- Hefetz, I.; Einot, N.; Faerman, M.; Horowitz, M.; Almog, J. Touch DNA: The Effect of the Deposition Pressure on the Quality of Latent Fingermarks and STR Profiles. Forensic Sci. Int. Genet. 2019, 38, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Währer, J.; Kehm, S.; Allen, M.; Brauer, L.; Eidam, O.; Seiberle, I.; Kron, S.; Scheurer, E.; Schulz, I. The DNA-Buster: The Evaluation of an Alternative DNA Recovery Approach. Forensic Sci. Int. Genet. 2023, 64, 102830. [Google Scholar] [CrossRef] [PubMed]
- Ostojic, L.; Wurmbach, E. Analysis of Fingerprint Samples, Testing Various Conditions, for Forensic DNA Identification. Sci. Justice 2017, 57, 35–40. [Google Scholar] [CrossRef] [PubMed]

| Glass | Plastic | |||
|---|---|---|---|---|
| Vac Dry | Vac Wet | Vac Dry | Vac Wet | |
| Chelex-Tween | 0.0002 | 0.0001 | 0.0001 | 0.0002 |
| 0.0004 | 0.0006 | 0.0021 | 0.003 | |
| 0.0002 | 0.0016 | 0.0062 | 0.0039 | |
| Month | Donor 001C (ng/µL) | Donor 002C (ng/µL) | Donor 005C (ng/µL) |
|---|---|---|---|
| 0 | 0.0019 | 0.0644 | 0.0003 |
| 1 | 0.0007 | 0.0183 | 0.0003 |
| 2 | 0.0033 | 3.2568 | 0.0014 |
| 3 | 0.0037 | 0.0248 | 0 |
| 4 | 0.0011 | 0.0301 | 0.001 |
| 5 | 0.0033 | 0.0861 | 0.0005 |
| 6 | 0.001 | 0.1125 | 0.0009 |
| 7 | 0.0002 | 0.0854 | 0.0017 |
| 8 | 0.0009 | 0.0413 | 0.0028 |
| 9 | 0.0021 | 0.0296 | 0.002 |
| 10 | 0.0012 | 0.0454 | 0.003 |
| 11 | 0.0009 | 0.0135 | 0.0007 |
| Average | 0.0016 | 0.0501 | 0.0012 |
| Standard Deviation | 0.0011 | 0.0324 | 0.0009 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morgan, A.G.; Prinz, M. Development of Improved DNA Collection and Extraction Methods for Handled Documents. Genes 2023, 14, 761. https://doi.org/10.3390/genes14030761
Morgan AG, Prinz M. Development of Improved DNA Collection and Extraction Methods for Handled Documents. Genes. 2023; 14(3):761. https://doi.org/10.3390/genes14030761
Chicago/Turabian StyleMorgan, Ashley G., and Mechthild Prinz. 2023. "Development of Improved DNA Collection and Extraction Methods for Handled Documents" Genes 14, no. 3: 761. https://doi.org/10.3390/genes14030761
APA StyleMorgan, A. G., & Prinz, M. (2023). Development of Improved DNA Collection and Extraction Methods for Handled Documents. Genes, 14(3), 761. https://doi.org/10.3390/genes14030761





