Evaluating the Effects of Cryopreservation on the Viability and Gene Expression of Porcine-Ear-Skin Fibroblasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Porcine-Ear-Skin-Sample Collection
2.2. Cell Cultures
2.3. Microbial Contamination Detection
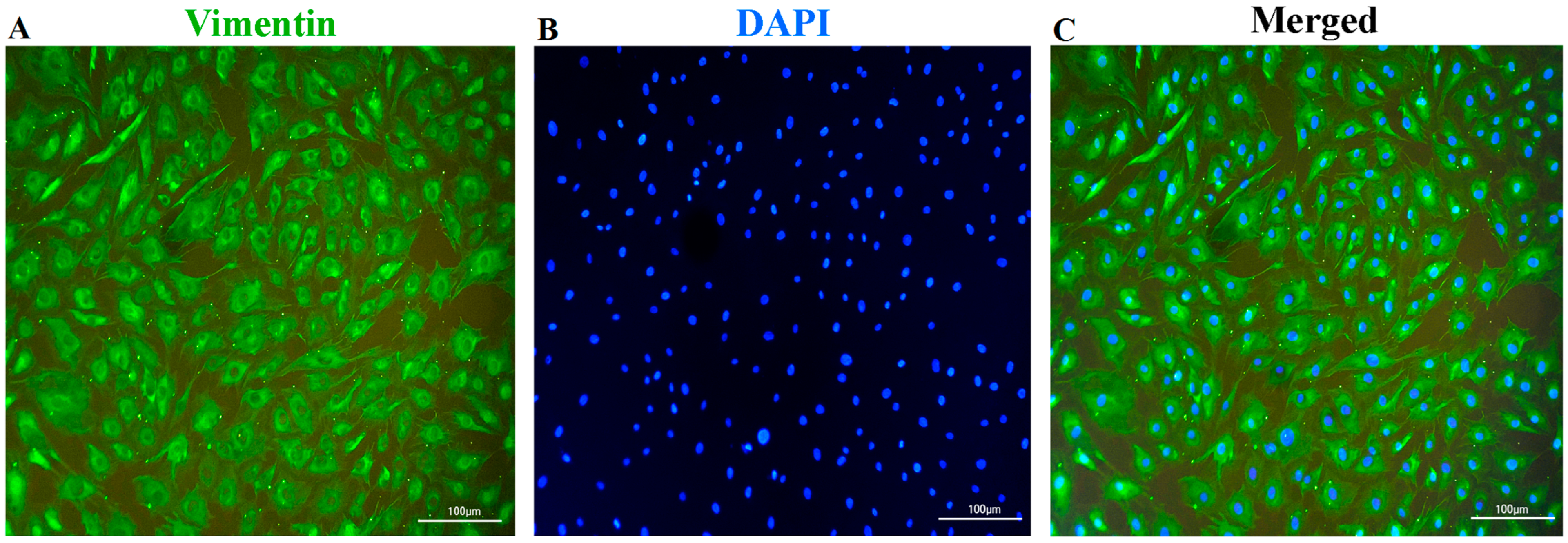
2.4. Identification of Fibroblasts
2.5. Cryopreservation, Recovery, and Cell-Viability Measurement
2.6. RNA-Sequencing Analysis
2.7. Quantitative Real-Time PCR Validation
2.8. Statistical Analysis
3. Results
3.1. Identification of the Fibroblasts and Cellular-Viability Measurement
3.2. RNA-Sequencing Data and Alignment-Quality Assessment
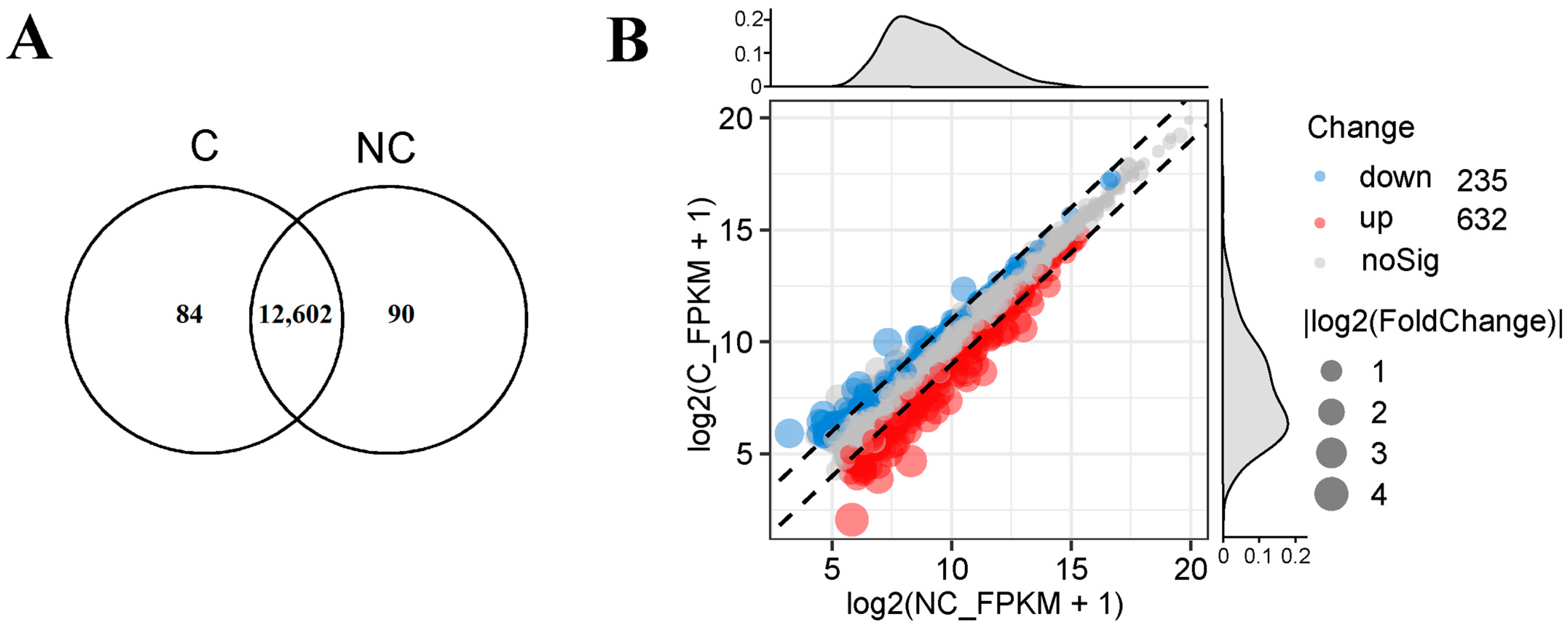
3.3. Differential Expression Genes (DEGs) of Fibroblasts in Response to Cryopreservation
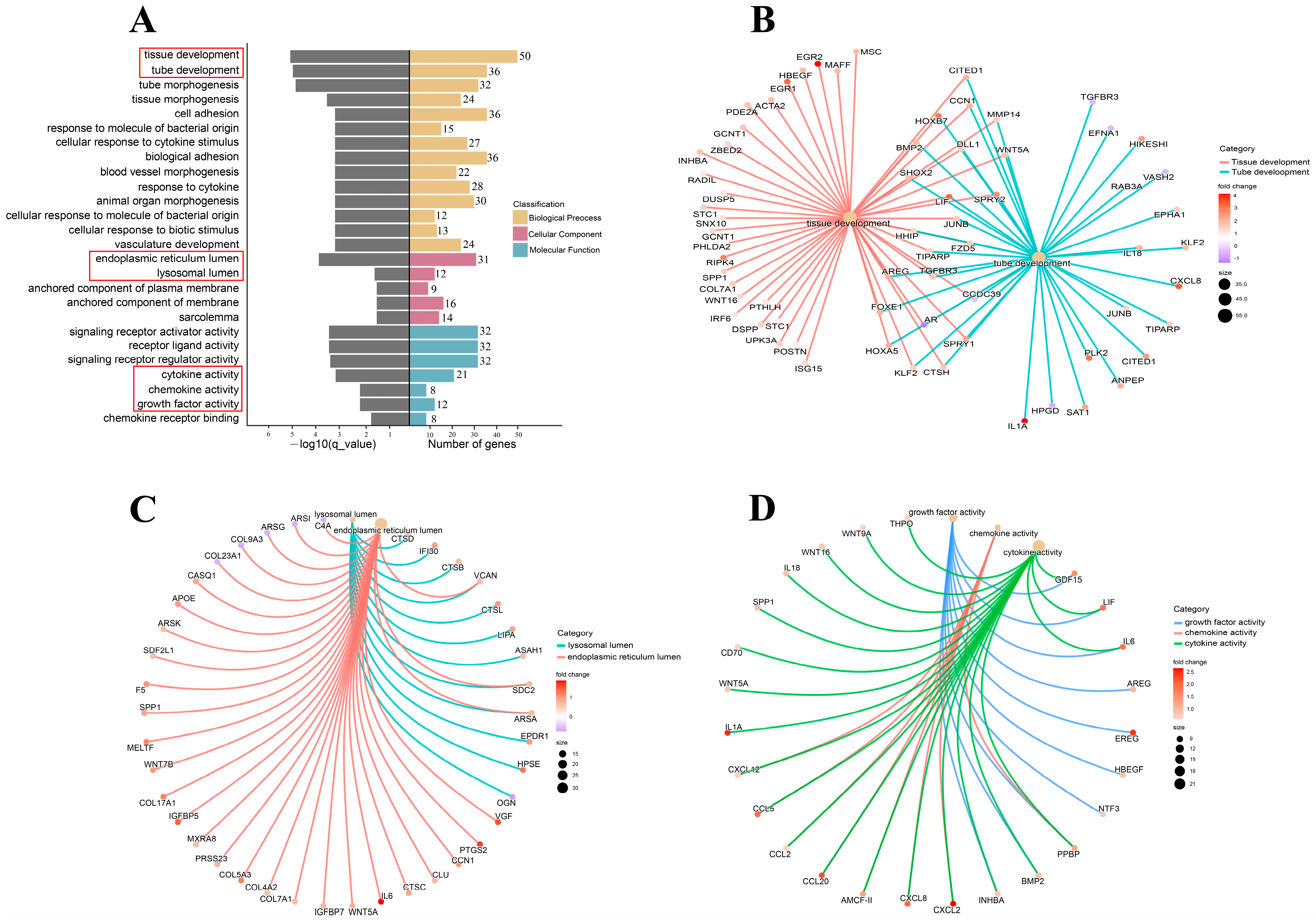
3.4. Cryopreservation-Induced Molecular-Functional Profile Alterations of Fibroblasts
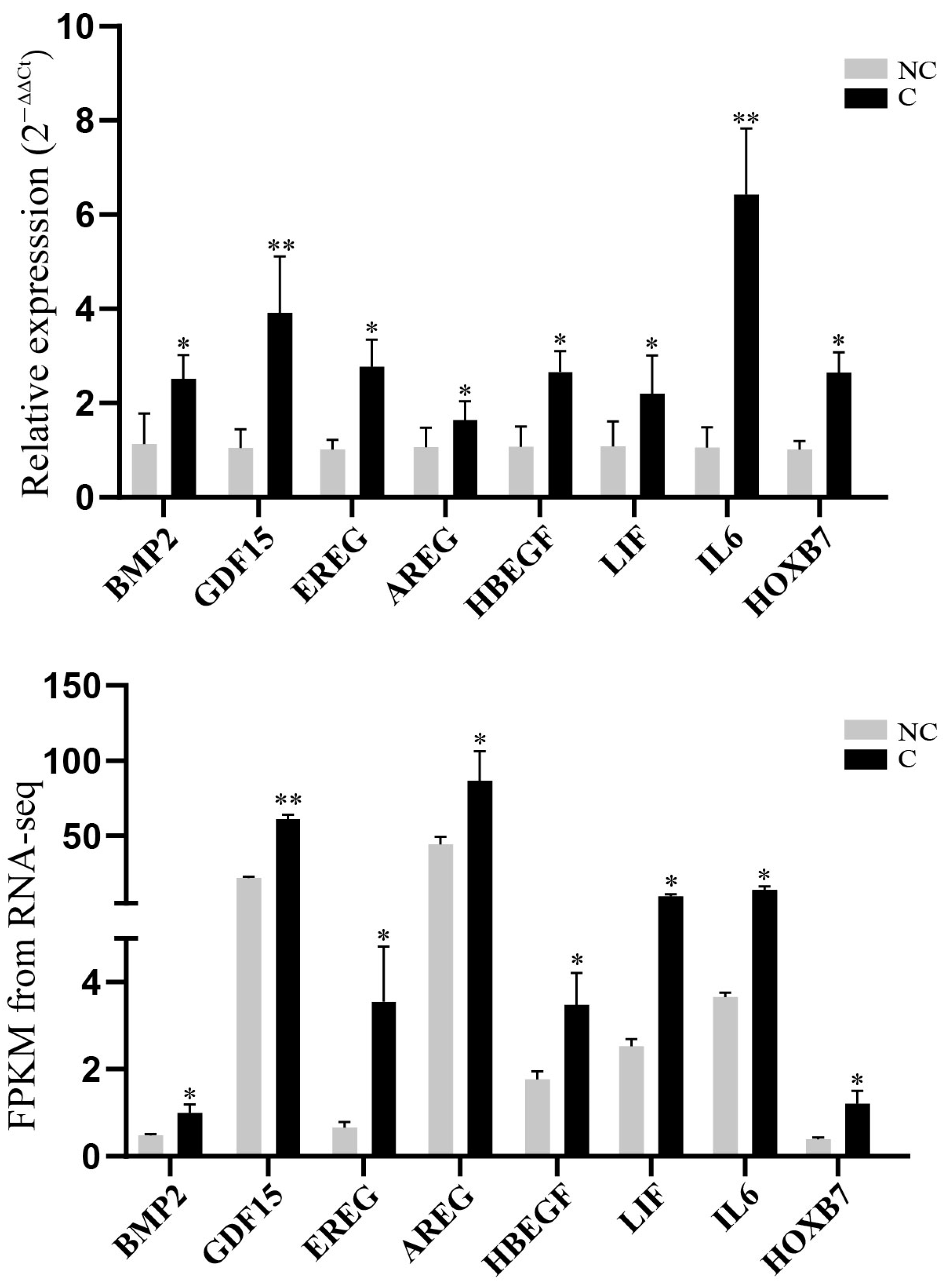
3.5. Validation of RNA-Sequencing Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plikus, M.V.; Wang, X.; Sinha, S.; Forte, E.; Thompson, S.M.; Herzog, E.L.; Driskell, R.R.; Rosenthal, N.; Biernaskie, J.; Horsley, V. Fibroblasts: Origins, definitions, and functions in health and disease. Cell 2021, 184, 3852–3872. [Google Scholar] [CrossRef] [PubMed]
- Kirk, T.; Ahmed, A.; Rognoni, E. Fibroblast Memory in Development, Homeostasis and Disease. Cells 2021, 10, 2480. [Google Scholar] [CrossRef] [PubMed]
- Madelaire, C.B.; Klink, A.C.; Israelsen, W.J.; Hindle, A.G. Fibroblasts as an experimental model system for the study of comparative physiology. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2022, 260, 110735. [Google Scholar] [CrossRef] [PubMed]
- Ujihira, M.; Iwama, A.; Aoki, M.; Aoki, K.; Omaki, S.; Goto, E.; Mabuchi, K. Cryoprotective effect of low-molecular-weight hyaluronan on human dermal fibroblast monolayers. CryoLetters 2010, 31, 101–111. [Google Scholar] [PubMed]
- Idda, A.; Bebbere, D.; Corona, G.; Masala, L.; Casula, E.; Cincotti, A.; Ledda, S. Insights on Cryopreserved Sheep Fibroblasts by Cryomicroscopy and Gene Expression Analysis. Biopreserv. Biobank 2017, 15, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Aydogdu, N.; Oztel, O.N.; Karaoz, E. Isolation, Culture, Cryopreservation, and Preparation of Skin-Derived Fibroblasts as a Final Cellular Product Under Good Manufacturing Practice-Compliant Conditions. Methods Mol. Biol. 2021, 2286, 85–94. [Google Scholar]
- Seluanov, A.; Vaidya, A.; Gorbunova, V. Establishing primary adult fibroblast cultures from rodents. J. Vis. Exp. 2010, 44, 2033. [Google Scholar]
- Bai, C.; Li, C.; Jin, D.; Guo, Y.; Guan, W.; Ma, Y.; Zhao, Q. Establishment and characterization of a fibroblast line from landrace. Artif. Cells Blood Substit. Biotechnol. 2010, 38, 129–135. [Google Scholar] [CrossRef]
- Praxedes, E.A.; Silva, M.B.; Oliveira, L.R.M.; Viana, J.; Silva, A.R.; Oliveira, M.F.; Pereira, A.F. Establishment, characterization, and cryopreservation of cell lines derived from red-rumped agouti (Dasyprocta leporina Linnaeus, 1758)—A study in a wild rodent. Cryobiology 2021, 98, 63–72. [Google Scholar] [CrossRef]
- Hamilton, D.W.; Walker, J.T.; Tinney, D.; Grynyshyn, M.; El-Warrak, A.; Truscott, E.; Flynn, L.E. The pig as a model system for investigating the recruitment and contribution of myofibroblasts in skin healing. Wound Repair Regen. 2022, 30, 45–63. [Google Scholar] [CrossRef]
- Zuo, Y.; Yi, L.; Lu, S. Dermal fibroblast from superficial layers of pig skin exhibits more proliferative capacity than that from deep layers. J. Tissue Viability 2022, 31, 278–285. [Google Scholar] [CrossRef]
- Li, L.; Pang, D.; Chen, L.; Wang, T.; Nie, D.; Yan, S.; Ouyang, H. Establishment of a transgenic pig fetal fibroblast reporter cell line for monitoring Cre recombinase activity. DNA Cell Biol. 2009, 28, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.F.; Muffley, L.A.; Seaton, M.E.; Ga, M.; Sirimahachaiyakul, P.; Hocking, A.M.; Gibran, N.S. Dermal Fibroblasts from the Red Duroc Pig Have an Inherently Fibrogenic Phenotype: An In Vitro Model of Fibroproliferative Scarring. Plast Reconstr. Surg. 2015, 136, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.J.; Crocini, C.; Ramirez, D.; Killaars, A.R.; Grim, J.C.; Aguado, B.A.; Clark, K.; Allen, M.A.; Dowell, R.D.; Leinwand, L.A.; et al. Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts. Nat. Biomed. Eng. 2021, 5, 1485–1499. [Google Scholar] [CrossRef]
- Smatlikova, P.; Askeland, G.; Vaskovicova, M.; Klima, J.; Motlik, J.; Eide, L.; Ellederova, Z. Age-Related Oxidative Changes in Primary Porcine Fibroblasts Expressing Mutated Huntingtin. Neurodegener. Dis. 2019, 19, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yuan, X.; Zhou, J.; Jin, J.; Zuo, Q.; Li, B. Comparison of the effects of three cryoprotectants on the cryopreservation of mouse subcutaneous tissue under different conditions. Exp. Ther. Med. 2020, 20, 3285–3289. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.A.S.; Borges, A.A.; Nascimento, M.B.; Aquino, L.V.C.; Silva, A.R.; Oliveira, M.F.; Pereira, A.F. Effects of Vitrification Techniques on the Somatic Tissue Preservation of Agouti (Dasyprocta leporina Linnaeus, 1758). Biopreserv. Biobank 2020, 18, 165–170. [Google Scholar] [CrossRef]
- Wang, T.; Li, Z.; Wei, J.; Zheng, D.; Wang, C.; Xu, C.; Chen, W.; Wang, B. Establishment and characterization of fibroblast cultures derived from a female common hippopotamus (Hippopotamus amphibius) skin biopsy. Cell Biol. Int. 2021, 45, 1571–1578. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Borges, A.A.; Lira, G.P.O.; Nascimento, L.E.; Santos, M.V.O.; Oliveira, M.F.; Silva, A.R.; Pereira, A.F. Isolation, characterization, and cryopreservation of collared peccary skin-derived fibroblast cell lines. PeerJ 2020, 8, e9136. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.B.; Praxedes, E.A.; Borges, A.A.; Oliveira, L.R.M.; Nascimento, M.B.; Silva, H.V.R.; Silva, A.R.; Pereira, A.F. Evaluation of the damage caused by in vitro culture and cryopreservation to dermal fibroblasts derived from jaguars: An approach to conservation through biobanks. Zoo Biol. 2021, 40, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Sass, K.; Corterier, C.; Brylla, E.; Loffler, S.; Steen, M.; Spanel-Borowski, K. Cryopreserved porcine tendons preserve cell viability after thawing. Transplant. Proc. 2009, 41, 1911–1913. [Google Scholar] [CrossRef]
- Fuller, B.J. Gene expression in response to low temperatures in mammalian cells: A review of current ideas. CryoLetters 2003, 24, 95–102. [Google Scholar]
- Singh, M.; Pierpoint, M.; Mikos, A.G.; Kasper, F.K. Chondrogenic differentiation of neonatal human dermal fibroblasts encapsulated in alginate beads with hydrostatic compression under hypoxic conditions in the presence of bone morphogenetic protein-2. J. Biomed. Mater. Res. A 2011, 98, 412–424. [Google Scholar] [CrossRef]
- Liu, Z.; Shen, J.; Pu, K.; Katus, H.A.; Ploger, F.; Tiefenbacher, C.P.; Chen, X.; Braun, T. GDF5 and BMP2 inhibit apoptosis via activation of BMPR2 and subsequent stabilization of XIAP. Biochim. Biophys. Acta 2009, 1793, 1819–1827. [Google Scholar] [CrossRef]
- Ding, L.Z.; Teng, X.; Zhang, Z.B.; Zheng, C.J.; Chen, S.H. Mangiferin inhibits apoptosis and oxidative stress via BMP2/Smad-1 signaling in dexamethasone-induced MC3T3-E1 cells. Int. J. Mol. Med. 2018, 41, 2517–2526. [Google Scholar] [CrossRef]
- Rochette, L.; Dogon, G.; Zeller, M.; Cottin, Y.; Vergely, C. GDF15 and Cardiac Cells: Current Concepts and New Insights. Int. J. Mol. Sci. 2021, 22, 8889. [Google Scholar] [CrossRef]
- Shrum, S.; Rusch, N.J.; MacMillan-Crow, L.A. Specific BK Channel Activator NS11021 Protects Rat Renal Proximal Tubular Cells from Cold Storage-Induced Mitochondrial Injury In Vitro. Biomolecules 2019, 9, 825. [Google Scholar] [CrossRef] [PubMed]
- Shrum, S.; Tobacyk, J.; Lo, S.; Parajuli, N.; MacMillan-Crow, L.A. The BK activator NS11021 partially protects rat kidneys from cold storage and transplantation-induced mitochondrial and renal injury. Arch. Biochem. Biophys. 2020, 688, 108410. [Google Scholar] [CrossRef] [PubMed]
- Golding, J.P.; Tsoni, S.; Dixon, M.; Yee, K.T.; Partridge, T.A.; Beauchamp, J.R.; Gassmann, M.; Zammit, P.S. Heparin-binding EGF-like growth factor shows transient left-right asymmetrical expression in mouse myotome pairs. Gene. Expr. Patterns 2004, 5, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Inatomi, O.; Andoh, A.; Yagi, Y.; Bamba, S.; Tsujikawa, T.; Fujiyama, Y. Regulation of amphiregulin and epiregulin expression in human colonic subepithelial myofibroblasts. Int. J. Mol. Med. 2006, 18, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Chu, E.K.; Foley, J.S.; Cheng, J.; Patel, A.S.; Drazen, J.M.; Tschumperlin, D.J. Bronchial epithelial compression regulates epidermal growth factor receptor family ligand expression in an autocrine manner. Am. J. Respir. Cell Mol. Biol. 2005, 32, 373–380. [Google Scholar] [CrossRef]
- Tschumperlin, D.J.; Shively, J.D.; Swartz, M.A.; Silverman, E.S.; Haley, K.J.; Raab, G.; Drazen, J.M. Bronchial epithelial compression regulates MAP kinase signaling and HB-EGF-like growth factor expression. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2002, 282, L904–L911. [Google Scholar] [CrossRef]
- Papaiahgari, S.; Yerrapureddy, A.; Hassoun, P.M.; Garcia, J.G.; Birukov, K.G.; Reddy, S.P. EGFR-activated signaling and actin remodeling regulate cyclic stretch-induced NRF2-ARE activation. Am. J. Respir. Cell Mol. Biol. 2007, 36, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Szabo, M.R.; Pipicz, M.; Csont, T.; Csonka, C. Modulatory Effect of Myokines on Reactive Oxygen Species in Ischemia/Reperfusion. Int. J. Mol. Sci. 2020, 21, 9382. [Google Scholar] [CrossRef]
- Yoshida, G.J. Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. J. Exp. Clin. Cancer Res. 2020, 39, 112. [Google Scholar] [CrossRef]
- Stoecklein, K.S.; Ortega, M.S.; Spate, L.D.; Murphy, C.N.; Prather, R.S. Improved cryopreservation of in vitro produced bovine embryos using FGF2, LIF, and IGF1. PLoS ONE 2021, 16, e0243727. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, Z.; Beer-Stolz, D.; Zimmers, T.A.; Koniaris, L.G. Interleukin-6 inhibits oxidative injury and necrosis after extreme liver resection. Hepatology 2007, 46, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.T.; Zhan, L.P.; Meng, C.; Zhang, N.; Chang, S.M.; Yao, R.; Li, C. Homeobox B7 promotes the osteogenic differentiation potential of mesenchymal stem cells by activating RUNX2 and transcript of BSP. Int. J. Clin. Exp. Med. 2015, 8, 10459–10470. [Google Scholar] [PubMed]
- Foppiani, E.M.; Candini, O.; Mastrolia, I.; Murgia, A.; Grisendi, G.; Samarelli, A.V.; Boscaini, G.; Pacchioni, L.; Pinelli, M.; De Santis, G.; et al. Impact of HOXB7 overexpression on human adipose-derived mesenchymal progenitors. Stem Cell Res. Ther. 2019, 10, 101. [Google Scholar] [CrossRef]
- Steens, J.; Unger, K.; Klar, L.; Neureiter, A.; Wieber, K.; Hess, J.; Jakob, H.G.; Klump, H.; Klein, D. Direct conversion of human fibroblasts into therapeutically active vascular wall-typical mesenchymal stem cells. Cell. Mol Life Sci. 2020, 77, 3401–3422. [Google Scholar] [CrossRef] [PubMed]
- Russo, B.; Borowczyk, J.; Boehncke, W.H.; Truchetet, M.E.; Modarressi, A.; Brembilla, N.C.; Chizzolini, C. Dysfunctional Keratinocytes Increase Dermal Inflammation in Systemic Sclerosis: Results From Studies Using Tissue-Engineered Scleroderma Epidermis. Arthritis Rheumatol. 2021, 73, 1311–1317. [Google Scholar] [CrossRef]
- Choi, J.; Bischof, J.C. Cooling rate dependent biophysical and viability response shift with attachment state in human dermal fibroblast cells. Cryobiology 2011, 63, 285–291. [Google Scholar] [CrossRef]


| Groups | Cryopreserved | Non-Cryopreserved |
|---|---|---|
| Total clean reads | 151,935,710 | 142,796,298 |
| Q30% | 94.56% | 94.30% |
| GC% | 52.26% | 52.26% |
| Mapped reads | 145,743,081 (95.92%) | 136,954,840 (95.90%) |
| Unique mapped reads | 140,741,266 (92.63%) | 132,860,276 (93.04%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Xie, Y.; Wang, J.; Huang, Y.; Zhang, X.; Xiao, T.; Fang, S. Evaluating the Effects of Cryopreservation on the Viability and Gene Expression of Porcine-Ear-Skin Fibroblasts. Genes 2023, 14, 751. https://doi.org/10.3390/genes14030751
Cao J, Xie Y, Wang J, Huang Y, Zhang X, Xiao T, Fang S. Evaluating the Effects of Cryopreservation on the Viability and Gene Expression of Porcine-Ear-Skin Fibroblasts. Genes. 2023; 14(3):751. https://doi.org/10.3390/genes14030751
Chicago/Turabian StyleCao, Jiacheng, Yingyu Xie, Jing Wang, Yongjie Huang, Xiaohan Zhang, Tianfang Xiao, and Shaoming Fang. 2023. "Evaluating the Effects of Cryopreservation on the Viability and Gene Expression of Porcine-Ear-Skin Fibroblasts" Genes 14, no. 3: 751. https://doi.org/10.3390/genes14030751
APA StyleCao, J., Xie, Y., Wang, J., Huang, Y., Zhang, X., Xiao, T., & Fang, S. (2023). Evaluating the Effects of Cryopreservation on the Viability and Gene Expression of Porcine-Ear-Skin Fibroblasts. Genes, 14(3), 751. https://doi.org/10.3390/genes14030751




