Polymorphisms and mRNA Expression Levels of IGF-1, FGF5, and KAP 1.4 in Tibetan Cashmere Goats
Abstract
1. Introduction
2. Materials and Methods
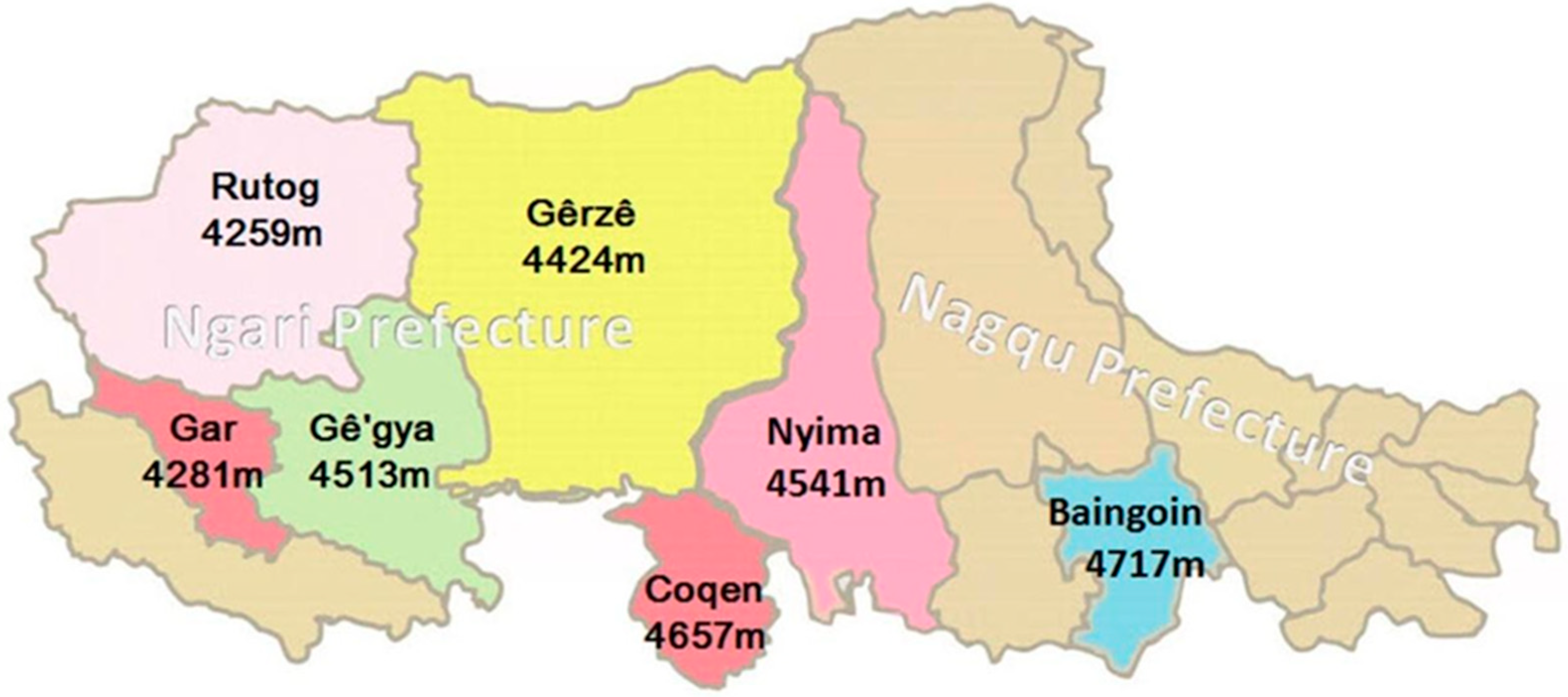
2.1. Experimental Animals
2.2. DNA and RNA Extraction and cDNA Synthesis
2.3. PCR and DNA Sequencing
2.4. Genotyping and Genetic Analysis
2.5. Quantitative PCR for mRNA Expression
2.6. Statistical Analysis
3. Results
3.1. Cloning of the Fragments of Genes IGF-1, FGF5, and KAP 1.4
3.2. Sequence Alignment and the Phylogenetic Tree
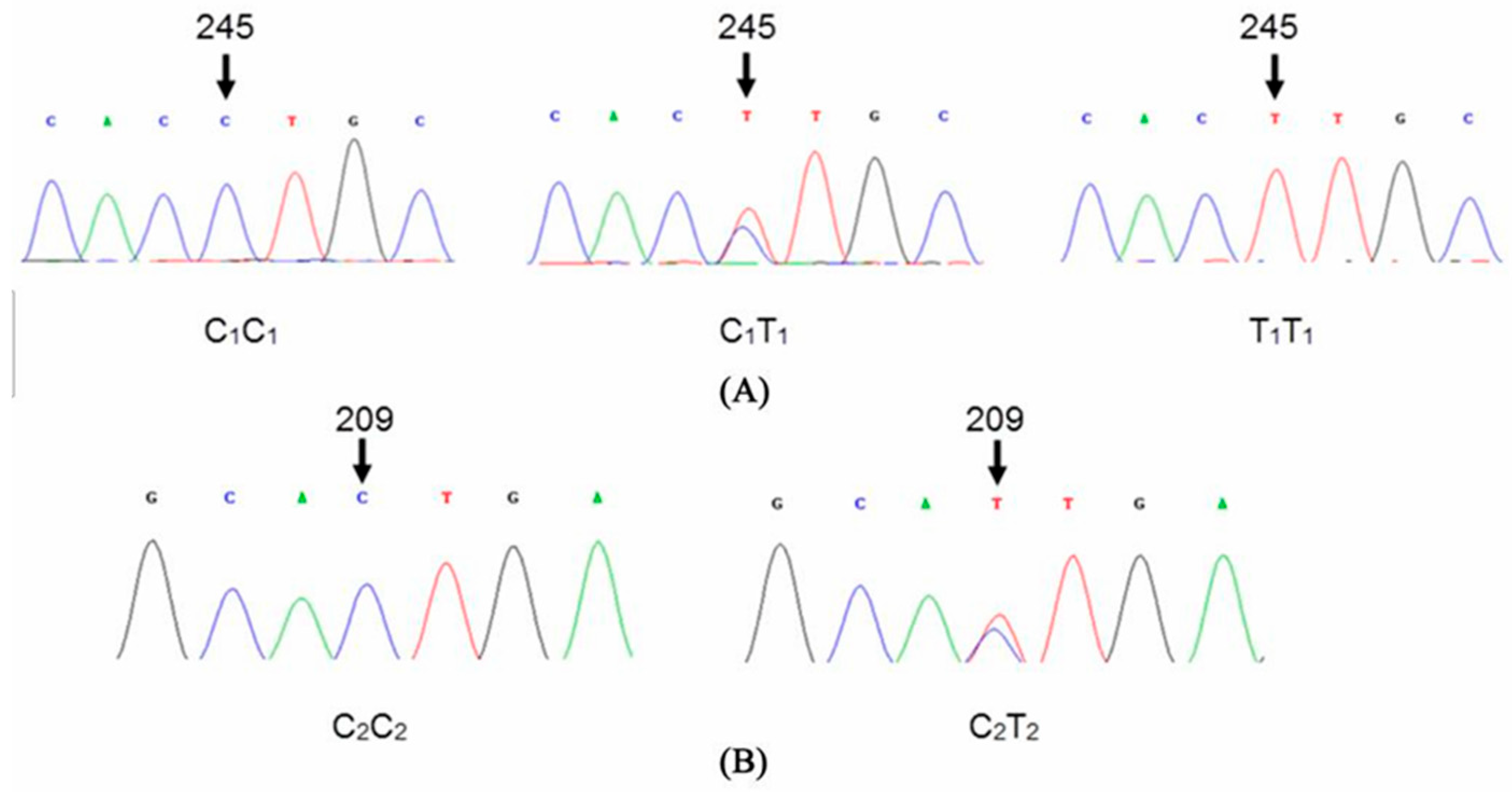
3.3. Single Nucleotide Polymorphism Analysis
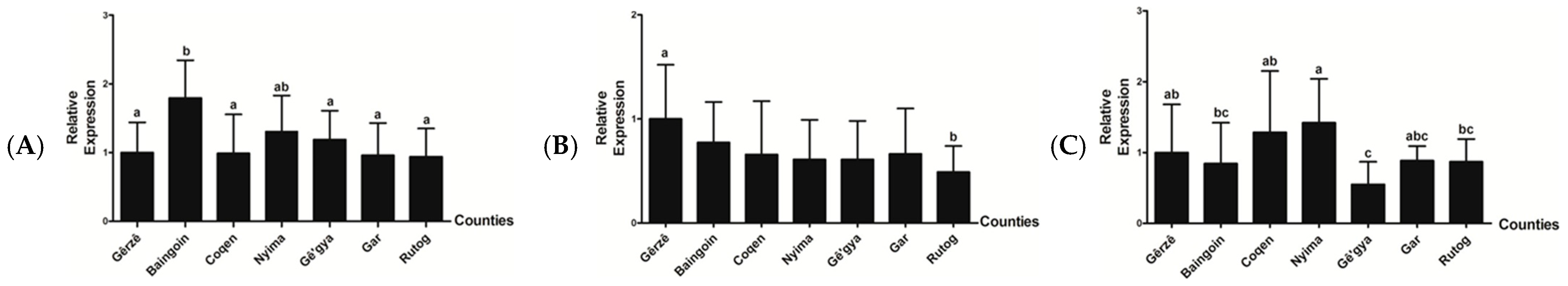
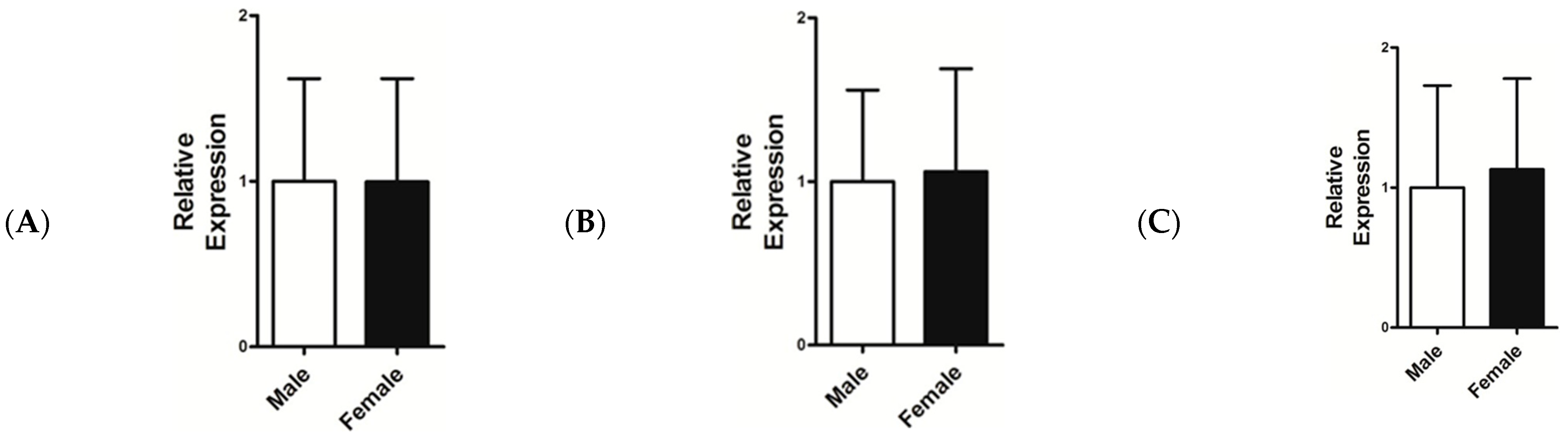
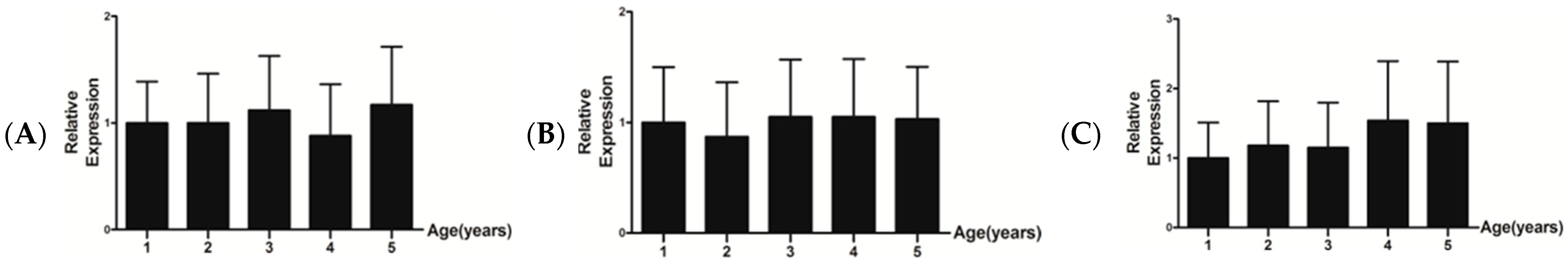
3.4. Relative mRNA Expression Level of IGF-1, FGF5, and KAP 1.4
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, Z.; Liu, Y.; He, X.; Di, R.; Wang, X.; Ren, C.; Zhang, Z.; Chu, M. Integrative Proteomics and Transcriptomics Profiles of the Oviduct Reveal the Prolificacy-Related Candidate Biomarkers of Goats (Capra hircus) in Estrous Periods. Int. J. Mol. Sci. 2022, 23, 14888. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Yao, N.; Yang, M.; Liu, X.; Dong, K.; Zhao, Q.; Pu, Y.; He, X.; Guan, W.; Yang, N.; et al. Exome sequencing reveals geneticdifferentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genom. 2016, 17, 1–12. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, W.; Song, W.; Sun, P.; Jia, Z. Effects of tryptophan supplementation on cashmere fiber characteristics, serum tryptophan, and related hormone concentrations in cashmere goats. Domest. Anim. Endocrinol. 2012, 43, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.L.; Dang, Y.L.; Yin, R.H.; Jiang, W.Q.; Wang, Z.Y.; Zhu, Y.B.; Wang, S.Q.; Zhao, Y.Y.; Deng, L.; Luo, G.B.; et al. Differential Expression of microRNAs and their Regulatory Networks in Skin Tissue of Liaoning Cashmere Goat during Hair Follicle Cycles. Anim. Biotechnol. 2016, 27, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Ansari-Renani, H.; Ebadi, Z.; Moradi, S.; Baghershah, H.; Ansari-Renani, M.; Ameli, S. Determination of hair follicle characteristics, density and activity of Iranian cashmere goat breeds. Small Rumin. Res. 2011, 95, 128–132. [Google Scholar] [CrossRef]
- Kurdistani, Z.K.; Rostamzadeh, J.; Rashidi, A.; Davis, M. Evaluation of insulin-like growth factor-I gene polymorphism on growth traits and yearling fleece weight in goats. Small Rumin. Res. 2013, 111, 10–15. [Google Scholar] [CrossRef]
- He, X.; Yuan, C.; Chen, Y. Isolation, characterization, and expression analysis of FGF5 isoforms in cashmere goat. Small Rumin. Res. 2014, 116, 111–117. [Google Scholar] [CrossRef]
- Shah, R.M.; Ganai, T.A.S.; Sheikh, F.D.; Shanaz, S.; Shabir, M.; Khan, H.M. Characterization and polymorphism of Keratin Associated Protein 1.4 gene in goats. Gene 2013, 518, 431–442. [Google Scholar] [CrossRef]
- Jin, M.; Liu, N.; Yuan, S.; Piao, J.; Zhao, F.; Qu, Y.; Zhang, T.; Wang, Y. Construction of a cDNA library and identification of genes from Liaoning cashmere goat. Livest. Sci. 2014, 164, 26–34. [Google Scholar] [CrossRef]
- Bai, W.L.; Yin, R.H.; Yin, R.L.; Wang, J.J.; Jiang, W.Q.; Luo, G.B.; Zhao, Z.H. IGF-1 mRNA splicing variants in Liaoning cashmere goat: Identification, characterization, and transcriptional patterns in skin and visceral organs. Anim. Biotechnol. 2013, 24, 81–93. [Google Scholar] [CrossRef]
- Liu, H.; Yang, G.; Zhang, W.; Zhu, X.; Jia, Z. Effects of FGF5 on fibre traits on Inner Mongolian cashmere goats. Hereditas 2009, 31, 175–179. (In Chinese) [Google Scholar]
- Wang, X.; Zhao, Z.D.; Xu, H.R.; Qu, L.; Zhao, H.B.; Li, T.; Zhang, Z.Y. Variation and expression of KAP 9.2 gene affecting cashmere trait in goats. Mol. Biol. Rep. 2012, 39, 10525–10529. [Google Scholar] [CrossRef]
- Thomas, N.; Venkatachalapathy, T.; Aravindakshan, T.; Raghavan, K.C. Molecular cloning, SNP detection and association analysis of 5′-flanking region of the goat IGF-1 gene with prolificacy. Anim. Reprod. Sci. 2016, 167, 8–15. [Google Scholar] [CrossRef]
- O’Sullivan, D.C.; Szestak, T.A.M.; Pell, J.M. Pell. Regulation of IGF-I mRNA by GH: Putative functions for class 1 and 2 message. Am. J. Physiol. Endocrinol. Metab. 2002, 283, 251–258. [Google Scholar] [CrossRef]
- Jansen, M.; van Schaik, F.M.A.; Ricker, A.T.; Bullock, B.; Woods, D.E.; Gabbay, K.H.; Nussbaum, A.L.; Sussenbach, J.S.; Van den Brande, J.L. Sequence of cDNA encoding human insulin-like growth factor I precursor. Nature 1983, 306, 609–611. [Google Scholar] [CrossRef]
- Ohlsen, S.M.; Dean, D.M.; Wong, E.A. Characterization of Multiple Transcription Initiation Sites of the Ovine Insulin-Like Growth Factor-I Gene and Expression Profiles of Three Alternatively Spliced Transcripts. DNA Cell Biol. 1993, 12, 3. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, W.; Luo, H.; Yue, W.; Gao, M.; Jia, Z. A New Single Nucleotide Polymorphism in the IGF-I Gene and Its Association with Growth Traits in the Nanjiang Huang Goat. Asian Aust. J. Anim. Sci. 2008, 21, 1073–1079. [Google Scholar] [CrossRef]
- He, X.; Chao, Y.; Zhou, G.; Chen, Y. Fibroblast growth factor 5-short(FGF5s) inhibits the activity of FGF5 in primary and secondary hair follicle dermal papilla cells of cashmere goats. Gene 2016, 575, 393–398. [Google Scholar] [CrossRef]
- Higgins, C.A.; Petukhova, L.; Harel, S.; Ho, Y.Y.; Drill, E.; Shapiro, L.; Wajid, M.; Christiano, A.M. FGF5 is a crucial regulator of hair length in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 10648–10653. [Google Scholar] [CrossRef]
- Hbbert, J.M.; Rosenquist, T.; Götz, J.; Martin, G.R. FGF5 as a regulator of the hair Growth Cycle: Evidence from Targeted adn Spontaneous Mutations. Cell 1994, 78, 1017–1205. [Google Scholar] [CrossRef]
- Sundberg, J.P.; Rourk, M.H.; Boggess, D.; Hogan, M.E.; Sundberg, B.A.; Bertolino, A.P. Angora Mouse Mutations: Altered Hair Cycle, Follicular Dystrophy, Phenotypic, Maintenance of Skin Grafts, and Changes in Keratin Expression. Vet. Pathol. 1997, 34, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Housley, D.J.E.; Venta, P.J. The long and the short of it: Evidence that FGF5 is a major determinant of canine “hair”-itability. Anim. Genet. 2006, 37, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Liu, W.J.; Zhang, F.Q.; Shao, Y.G.; Yu, S.G. The polymorphism of a Novel Mutation of KAP 13.1 Gene and its Association with Cashmere Traits on Xinjiang Local Goat Breed in China. Asian J. Anim. Vet. Adv. 2010, 5, 34–42. [Google Scholar] [CrossRef]
- Parsons, Y.M.; Cooper, D.W.; Piper, L.R. Evidence of linkage between high-glycine-tyrosine keratin gene loci and wool fibre diameter in a Merino half-sib family. Anim. Genet. 1994, 25, 105–108. [Google Scholar] [CrossRef]
- Gao, A.I.; Li, N.; Zhao, X. PCR-SSCP Analysis of FGF5 Gene inDifferent Sheep Breeds. Acta Vet. Et Zootech. Sin. 2006, 37, 326. (In Chinese) [Google Scholar]
- Vahidi, S.M.F.; Tarang, A.R.; Naqvi, A.-U.; Anbaran, M.F.; Boettcher, P.; Joost, S.; Colli, L.; Garcia, J.F.; Ajmone-Marsan, P. Investigation of the genetic diversity of domestic Capra hircus breeds reared within an early goat domestication area in Iran. Genet. Sel. Evol. 2014, 46, 1–12. [Google Scholar] [CrossRef]
- Rosenquist, T.A.; Martin, G.R. Fibroblast Growth Factor Signalling in the Hair Growth Cycle: Expression of the Fibroblast Growth Factor Receptor and Ligand Genes in the Murine Hair Follicle. Dev. Dyn. 1996, 205, 379–386. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Hickford, J.G.H. Polymorphism of the ovine keratin-associated protein 1–4 gene (KRTAP1.4). Mol. Biol. Rep. 2010, 37, 3377–3380. [Google Scholar] [CrossRef]
- Brito, L.F.; Jafarikia, M.; Grossi, D.A.; Kijas, J.W.; Porto-Neto, L.R.; Ventura, R.V.; Salgorzaei, M.; Schenkel, F.S. Characterization of linkage disequilibrium, consistency of gametic phase and admixture in Australian and Canadian goats. BMC Genet. 2015, 16, 1–15. [Google Scholar] [CrossRef]
- Wang, J.; Hao, Z.; Zhou, H.; Luo, Y.; Hu, J.; Liu, X.; Li, S.; Hickford, J.G. A keratin-associated protein (KAP) gene that is associated with variation in cashmere goat fleece weight. Small Rumin. Res. 2018, 167, 104–109. [Google Scholar] [CrossRef]
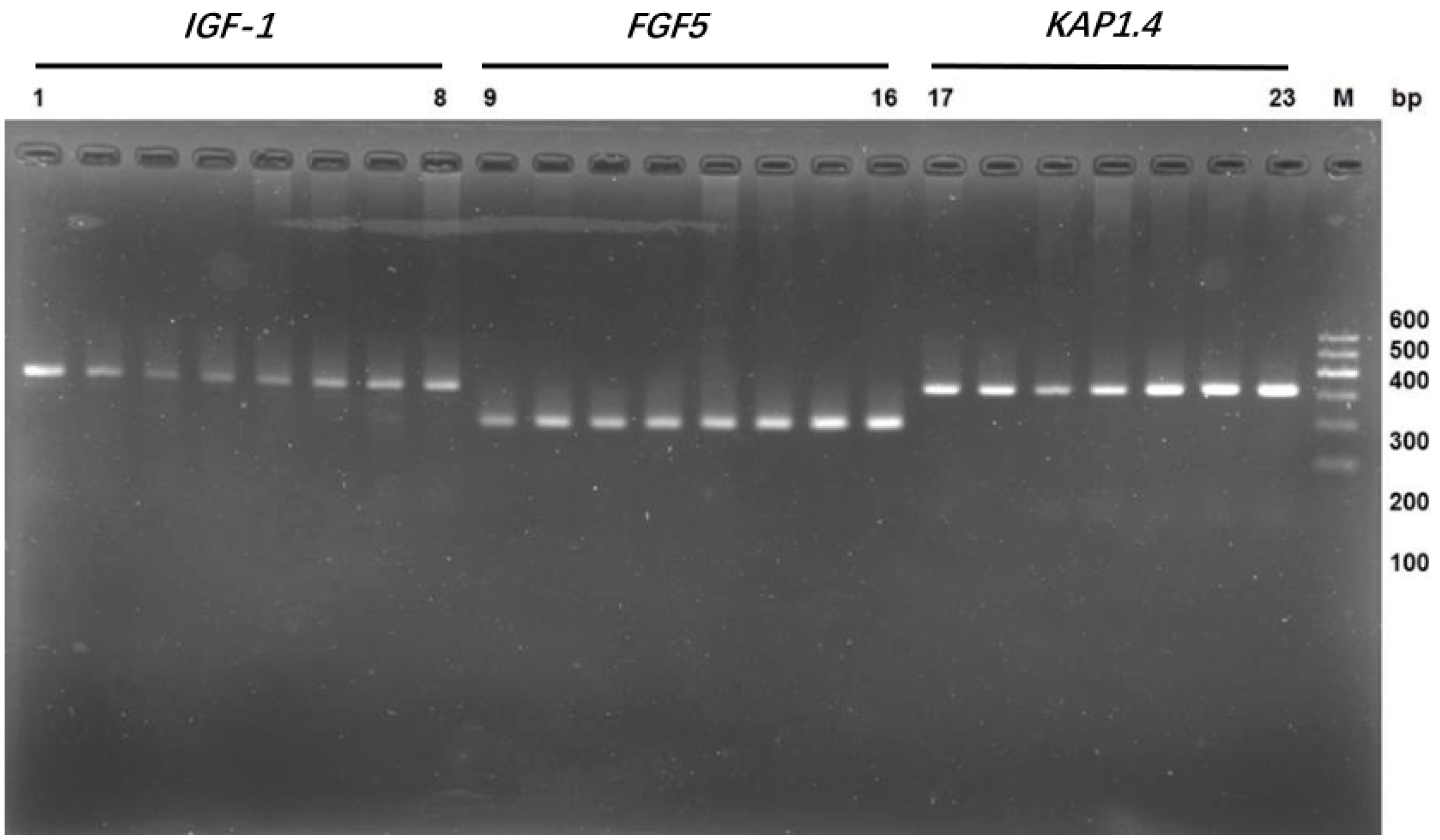

| Primer Name | Primer Sequence (5′-3′) | Product Size (bp) | Annealing Temperature (°C) | Accession Number |
|---|---|---|---|---|
| Primers for SNP detection | ||||
| IGF1-F | GCTGGGTGTAGCAGTGAACA | 320 | 55.6 | D26119 |
| IGF1-R | GTTGCTTCAGCCGCATAACT | |||
| FGF5-F | AGCAGTAGCACCGTGTCTTC | 193 | 61.0 | XM_01294679 |
| FGF5-R | AGCCATTGACTTTGCCATCC | |||
| KAP1.4-F | TTGGTGGCAGCATTGGCTATG | 303 | 54.7 | GQ507748 |
| KAP1.4-R | AGAGATACTGTGTCTTGGGCA | |||
| Primers for real time PCR | ||||
| IGF1-F | AATCAGCAGTCTTCCAACCCAA | 114 | 51.8 | NM_001285697 |
| IGF1-R | AGCAAGCACAGGGCCAGAT | |||
| FGF5-F | ACCTCAGCACGTCTCTACCCAC | 123 | 52.4 | KM_596772 |
| FGF5-R | GGAACCTTTGGCTTGACGG | |||
| KAP1.4-F | GCCAGCCAACTTCCATCCA | 110 | 56.7 | JQ627657 |
| KAP1.4-R | AATGCCACAGCCGGTCTCAC | |||
| β-action-F | GGCCGCACCACTGGCATTGTCAT | 104 | 60.0 | DQ845171.1 |
| β-action-R | AGGTCCAGACGCAGGATGGCG | |||
| Gene | Position of Mutation | Base Change | Codon | Amino Acid |
|---|---|---|---|---|
| KAP1.4 | 209 | C–T | C*TG–TTG | Leucine(L)–non-change |
| 245 | C–T | CC*T–CTT | Proline(P)–Leucine(L) |
| Counties | Genotypes | Frequencies | Alleles | Frequencies | Heterozygosity | Homozygosity |
|---|---|---|---|---|---|---|
| C1C1 | 0.86 | C1 | 0.930 | |||
| Gar (N = 37) | C1T1 | 0.14 | T1 | 0.070 | 0.135 | 0.865 |
| T1T1 | 0 | |||||
| C1C1 | 0.84 | C1 | 0.895 | |||
| Rutog (N = 37) | C1T1 | 0.11 | T1 | 0.105 | 0.108 | 0.892 |
| T1T1 | 0.05 | |||||
| C1C1 | 0.73 | C1 | 0.865 | |||
| Gê’gya (N = 30) | C1T1 | 0.27 | T1 | 0.135 | 0.266 | 0.734 |
| T1T1 | 0 | |||||
| C1C1 | 0.78 | C1 | 0.890 | |||
| Baingoin (N = 37) | C1T1 | 0.22 | T1 | 0.110 | 0.216 | 0.784 |
| T1T1 | 0 | |||||
| C1C1 | 0.73 | C1 | 0.845 | |||
| Gêrzê (N = 30) | C1T1 | 0.23 | T1 | 0.155 | 0.233 | 0.767 |
| T1T1 | 0.04 | |||||
| C1C1 | 0.57 | C1 | 0.755 | |||
| Coqen (N = 35) | C1T1 | 0.37 | T1 | 0.245 | 0.371 | 0.629 |
| T1T1 | 0.06 | |||||
| C1C1 (N = 156) | 0.76 | C1 | 0.870 | |||
| Total (N = 206) Nyima C1T1 (N = 45) | 0.22 | T1 | 0.130 | 0.218 | 0.782 | |
| T1T1 (N = 5) | 0.02 | |||||
| Genetic Group | Genotypes | Frequencies | Alleles | Frequencies | Heterozygosity | Homozygosity |
|---|---|---|---|---|---|---|
| C2C2 | 0.97 | C2 | 0.985 | |||
| Gar (N = 37) | C2T2 | 0.03 | T2 | 0.015 | 0.027 | 0.973 |
| T2T2 | 0 | |||||
| C2C2 | 0.92 | C2 | 0.960 | |||
| Rutog (N = 37) | C2T2 | 0.08 | T2 | 0.040 | 0.081 | 0.919 |
| T2T2 | 0 | |||||
| C2C2 | 0.93 | C2 | 0.965 | |||
| Gê’gya (N = 30) | C2T2 | 0.07 | T2 | 0.035 | 0.066 | 0.934 |
| T2T2 | 0 | |||||
| C2C2 | 1 | C2 | 1.000 | |||
| Baingoin (N = 37) | C2T2 | 0 | T2 | 0.000 | 0.000 | 1.000 |
| T2T2 | 0 | |||||
| C2C2 | 0.93 | C2 | 0.965 | |||
| Gêrzê (N = 30) | C2T2 | 0.07 | T2 | 0.035 | 0.066 | 0.934 |
| T2T2 | 0 | |||||
| C2C2 | 0.89 | C2 | 0.945 | |||
| Coqen (N = 35) | C2T2 | 0.11 | T2 | 0.055 | 0.114 | 0.886 |
| T2T2 | 0 | |||||
| C2C2 (N = 194) | 0.94 | C2 | 0.970 | |||
| Total (N = 206) | C2T2 (N = 12) | 0.06 | T2 | 0.030 | 0.058 | 0.942 |
| T2T2 (N = 0) | 0 | |||||
| Counties | Rutog | Coqen | Nyima | Baingoin | Gê’gya | Gêrzê | Gar |
|---|---|---|---|---|---|---|---|
| Average cashmere production (g/each) | 380 | 350 | 350 | 300 | 300 | 300 | 300 |
| Age | 1-Year-Old | 2-Year-Old | 3-Year-Old | 4-Year-Old | 5-Year-Old |
|---|---|---|---|---|---|
| Average cashmere production (g/each) | <50 | 50–200 | 50–200 | 50–200 | 50–200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, T.; Tan, Y.; Cuomu, R.; Liu, Y.; Ba, G.; Suo, L.; Wu, Y.; Cao, X.; Zeng, X. Polymorphisms and mRNA Expression Levels of IGF-1, FGF5, and KAP 1.4 in Tibetan Cashmere Goats. Genes 2023, 14, 711. https://doi.org/10.3390/genes14030711
Song T, Tan Y, Cuomu R, Liu Y, Ba G, Suo L, Wu Y, Cao X, Zeng X. Polymorphisms and mRNA Expression Levels of IGF-1, FGF5, and KAP 1.4 in Tibetan Cashmere Goats. Genes. 2023; 14(3):711. https://doi.org/10.3390/genes14030711
Chicago/Turabian StyleSong, Tianzeng, Yao Tan, Renqing Cuomu, Yacheng Liu, Gui Ba, Langda Suo, Yujiang Wu, Xiaohan Cao, and Xianyin Zeng. 2023. "Polymorphisms and mRNA Expression Levels of IGF-1, FGF5, and KAP 1.4 in Tibetan Cashmere Goats" Genes 14, no. 3: 711. https://doi.org/10.3390/genes14030711
APA StyleSong, T., Tan, Y., Cuomu, R., Liu, Y., Ba, G., Suo, L., Wu, Y., Cao, X., & Zeng, X. (2023). Polymorphisms and mRNA Expression Levels of IGF-1, FGF5, and KAP 1.4 in Tibetan Cashmere Goats. Genes, 14(3), 711. https://doi.org/10.3390/genes14030711




